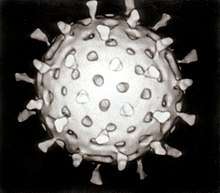
Orthomyxoviridae
Orthomyxoviridae (ὀρθός, orthós, Greek for "straight"; μύξα, mýxa, Greek for "mucus")[1] is a family of RNA viruses. It includes seven genera: Influenzavirus A, Influenzavirus B, Influenzavirus C, Influenzavirus D, Isavirus, Thogotovirus, and Quaranjavirus. The first four genera contain viruses that cause influenza in vertebrates, including birds (see also avian influenza), humans, and other mammals. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates, such as ticks and mosquitoes.[2][3][4]
| Orthomyxovirus | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Phylum: | Negarnaviricota |
| Class: | Insthoviricetes |
| Order: | Articulavirales |
| Family: | Orthomyxoviridae |
| Genera | |
| |
| Influenza (Flu) |
|---|
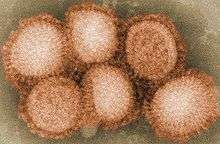 |
| Types |
|
| Vaccines |
|
| Treatment |
| Pandemics |
|
| Outbreaks |
|
| See also |
|
The four genera of Influenza virus, which are identified by antigenic differences in their nucleoprotein and matrix protein, infect vertebrates as follows:
- Influenzavirus A infects humans, other mammals, and birds, and causes all flu pandemics
- Influenzavirus B infects humans and seals
- Influenzavirus C infects humans, pigs, and dogs.
- Influenzavirus D infects pigs and cattle
Classification
In a phylogenetic-based taxonomy, the category "RNA virus" includes the category "negative-sense ssRNA virus", which includes the Order "Mononegavirales", and the Family "Orthomyxovirus" (among others). The genera-associated species and serotypes of Orthomyxovirus are shown in the following table.
| Genus | Species (* indicates type species) | Serotypes or Subtypes | Hosts |
|---|---|---|---|
| Influenza virus A | Influenza A virus* | H1N1, H1N2, H2N2, H3N1, H3N2, H3N8, H5N1, H5N2, H5N3, H5N8, H5N9, H7N1, H7N2, H7N3, H7N4, H7N7, H7N9, H9N2, H10N7 | Human, pig, bird, horse, bat |
| Influenza virus B | Influenza B virus* | Victoria, Yamagata[5] | Human, seal |
| Influenza virus C | Influenza C virus* | Human, pig, dog | |
| Influenza virus D | Influenza D virus* | Pig, cattle | |
| Isavirus | Infectious salmon anemia virus* | Atlantic salmon | |
| Thogotovirus | Thogotovirus* | Tick, mosquito, mammal (including human) | |
| Dhori virus | Batken virus, Bourbon virus, Jos virus | ||
| Quaranjavirus[6] | |||
| Quaranfil virus,* Johnston Atoll virus | |||
ICTV Taxonomy
Group: ssRNA(-)
- Family: Orthomyxoviridae
- Genus: Influenzavirus C
- Influenza C virus
- Genus: Isavirus
- Infectious salmon anemia virus
- Genus: Quaranjavirus
- Johnston Atoll virus
- Quaranfil virus
- Genus: Thogotovirus
- Dhori virus
- Thogoto virus
Types
There are four genera of influenza virus, each containing only a single species, or type. Influenza A and C infect a variety of species, while influenza B almost exclusively infects humans, and influenza D infects cattle and pigs.[8][9][10]
Influenza A
Influenza A viruses are further classified, based on the viral surface proteins hemagglutinin (HA or H) and neuraminidase (NA or N). Sixteen H subtypes (or serotypes) and nine N subtypes of influenza A virus have been identified.
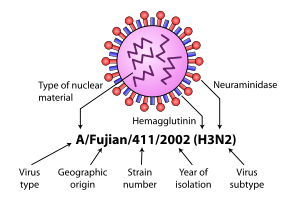
Further variation exists; thus, specific influenza strain isolates are identified by a standard nomenclature specifying virus type, geographical location where first isolated, sequential number of isolation, year of isolation, and HA and NA subtype.[11][12]
Examples of the nomenclature are:
- A/Brisbane/59/2007 (H1N1)
- A/Moscow/10/99 (H3N2).
The type A viruses are the most virulent human pathogens among the three influenza types and cause the most severe disease. The serotypes that have been confirmed in humans, ordered by the number of known human pandemic deaths, are:
- H1N1 caused "Spanish flu" in 1918, "Swine flu" in 2009.[13]
- H2N2 caused "Asian Flu".
- H3N2 caused "Hong Kong Flu".
- H5N1 is a pandemic threat.[14]
- H7N7 has unusual zoonotic potential.[15]
- H1N2 is endemic in humans and pigs .
- H9N2, H7N2, H7N3, H10N7.
| Name of pandemic | Date | Deaths | Case fatality rate | Subtype involved | Pandemic Severity Index |
|---|---|---|---|---|---|
| 1889–1890 flu pandemic (Asiatic or Russian Flu)[19] |
1889–1890 | 1 million | 0.15% | possibly H3N8 or H2N2 | NA |
| 1918 flu pandemic (Spanish flu)[20] |
1918–1920 | 20 to 100 million | 2% | H1N1 | 5 |
| Asian Flu | 1957–1958 | 1 to 1.5 million | 0.13% | H2N2 | 2 |
| Hong Kong Flu | 1968–1969 | 0.75 to 1 million | <0.1% | H3N2 | 2 |
| Russian flu | 1977–1978 | no accurate count | N/A | H1N1 | N/A |
| 2009 flu pandemic[21][22] | 2009–2010 | 105,700–395,600[23] | 0.03% | H1N1 | NA |
Influenza B
Influenza B virus is almost exclusively a human pathogen, and is less common than influenza A. The only other animal known to be susceptible to influenza B infection is the seal.[24] This type of influenza mutates at a rate 2–3 times lower than type A[25] and consequently is less genetically diverse, with only one influenza B serotype.[8] As a result of this lack of antigenic diversity, a degree of immunity to influenza B is usually acquired at an early age. However, influenza B mutates enough that lasting immunity is not possible.[26] This reduced rate of antigenic change, combined with its limited host range (inhibiting cross species antigenic shift), ensures that pandemics of influenza B do not occur.[27]
Influenza C
The influenza C virus infects humans and pigs, and can cause severe illness and local epidemics.[28] However, influenza C is less common than the other types and usually seems to cause mild disease in children.[29][30]
Influenza D
This is a genus that was classified in 2016, the members of which were first isolated in 2011.[31] This genus appears to be most closely related to Influenza C, from which it diverged several hundred years ago.[32] There are at least two strains of this genus in extant.[33] The main hosts appear to be cattle, but this virus has seen to infect pigs as well.
Morphology
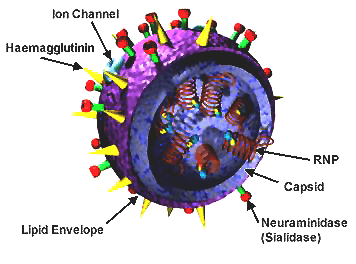
The virion is pleomorphic; the envelope can occur in spherical and filamentous forms. In general, the virus's morphology is ellipsoidal with particles 80 to 120 nm in diameter, or filamentous virions 80–120 nm in diameter and up to 20 µm long.[34] There are some 500 distinct spike-like surface projections of the envelope each projecting 10 to 14 nm from the surface with varying surface densities.
The major glycoprotein (HA) is interposed irregularly by clusters of neuraminidase (NA), with a ratio of HA to NA of about 4–5 to 1.
Cholesterol-laden membranes with protruding glycoproteins enclose the nucleocapsids; nucleoproteins of different size classes with a loop at each end; the arrangement within the virion is uncertain. The ribonuclear proteins are filamentous and fall in the range of 50 to 130 nm long and 9 to 15 nm in diameter. They have a helical symmetry.
Genome
Viruses of this family contain 6 to 8 segments of linear negative-sense single stranded RNA.[35]
The total genome length is 12000–15000 nucleotides (nt). The size of each segment is as follows:
| segment | protein | size (nt) | protein size (aa) |
|---|---|---|---|
| PB1 | polymerase | 2300–2500 | 757+87 (F2) |
| PB2 | polymerase | 2300–2500 | 759 |
| PA | polymerase | 2200–2300 | 716 |
| HA | Hemagglutinin | 1700–1800 | 550 |
| NP | nucleoprotein | 1500–1600 | 498 |
| NA | Neuraminidase | 1400–1500 | 454 |
| M | Membrane protein(s) | 1000–1100 | 252+97 |
| NS | non-structural protein(s) | 800–900 | 230+121 |
The Genome sequence has terminal repeated sequences; repeated at both ends. Terminal repeats at the 5'-end 12–13 nucleotides long. Nucleotide sequences of 3'-terminus identical; the same in genera of same family; most on RNA (segments), or on all RNA species. Terminal repeats at the 3'-end 9–11 nucleotides long. Encapsidated nucleic acid is solely genomic. Each virion may contain defective interfering copies. In Influenza A (H1N1) PB1-F2 is produced from an alternative reading frame in PB1. The M and NS genes produce 2 different genes via alternative splicing.[36]
Structure
The following applies for Influenza A viruses, although other influenza strains are very similar in structure:[37]
The influenza A virus particle or virion is 80–120 nm in diameter, usually producing both ellipsoidal, baciliform, and filamentous particles.[38][39][40] Unusually for a virus, the influenza A genome is not a single piece of nucleic acid; instead, it contains eight pieces of segmented negative-sense RNA (13.5 kilobases total), which encode 11 proteins (HA, NA, NP, M1, M2, NS1, NEP, PA, PB1, PB1-F2, PB2).[41] The best-characterised of these viral proteins are hemagglutinin and neuraminidase, two large glycoproteins found on the outside of the viral particles. Neuraminidase is an enzyme involved in the release of progeny virus from infected cells, by cleaving sugars that bind the mature viral particles. By contrast, hemagglutinin is a lectin that mediates binding of the virus to target cells and entry of the viral genome into the target cell.[42] The hemagglutinin (H) and neuraminidase (N) proteins are targets for antiviral drugs.[43] These proteins are also recognised by antibodies, i.e. they are antigens.[16] The responses of antibodies to these proteins are used to classify the different serotypes of influenza A viruses, hence the H and N in H5N1.
Replication cycle
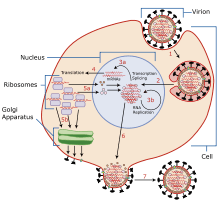
Typically, influenza is transmitted from infected mammals through the air by coughs or sneezes, creating aerosols containing the virus, and from infected birds through their droppings. Influenza can also be transmitted by saliva, nasal secretions, feces and blood. Infections occur through contact with these bodily fluids or with contaminated surfaces. Out of a host, flu viruses can remain infectious for about one week at human body temperature, over 30 days at 0 °C (32 °F), and indefinitely at very low temperatures (such as lakes in northeast Siberia). They can be inactivated easily by disinfectants and detergents.[44][45][46]
The viruses bind to a cell through interactions between its hemagglutinin glycoprotein and sialic acid sugars on the surfaces of epithelial cells in the lung and throat (Stage 1 in infection figure).[47] The cell imports the virus by endocytosis. In the acidic endosome, part of the haemagglutinin protein fuses the viral envelope with the vacuole's membrane, releasing the viral RNA (vRNA) molecules, accessory proteins and RNA-dependent RNA polymerase into the cytoplasm (Stage 2).[48] These proteins and vRNA form a complex that is transported into the cell nucleus, where the RNA-dependent RNA polymerase begins transcribing complementary positive-sense cRNA (Steps 3a and b).[49] The cRNA is either exported into the cytoplasm and translated (step 4), or remains in the nucleus. Newly synthesised viral proteins are either secreted through the Golgi apparatus onto the cell surface (in the case of neuraminidase and hemagglutinin, step 5b) or transported back into the nucleus to bind vRNA and form new viral genome particles (step 5a). Other viral proteins have multiple actions in the host cell, including degrading cellular mRNA and using the released nucleotides for vRNA synthesis and also inhibiting translation of host-cell mRNAs.[50]
Negative-sense vRNAs that form the genomes of future viruses, RNA-dependent RNA transcriptase, and other viral proteins are assembled into a virion. Hemagglutinin and neuraminidase molecules cluster into a bulge in the cell membrane. The vRNA and viral core proteins leave the nucleus and enter this membrane protrusion (step 6). The mature virus buds off from the cell in a sphere of host phospholipid membrane, acquiring hemagglutinin and neuraminidase with this membrane coat (step 7).[51] As before, the viruses adhere to the cell through hemagglutinin; the mature viruses detach once their neuraminidase has cleaved sialic acid residues from the host cell.[47] After the release of new influenza virus, the host cell dies.
Orthomyxoviridae viruses are one of two RNA viruses that replicate in the nucleus (the other being retroviridae). This is because the machinery of orthomyxo viruses cannot make their own mRNAs. They use cellular RNAs as primers for initiating the viral mRNA synthesis in a process known as cap snatching.[52] Once in the nucleus, the RNA Polymerase Protein PB2 finds a cellular pre-mRNA and binds to its 5' capped end. Then RNA Polymerase PA cleaves off the cellular mRNA near the 5' end and uses this capped fragment as a primer for transcribing the rest of the viral RNA genome in viral mRNA.[53] This is due to the need of mRNA to have a 5' cap in order to be recognized by the cell's ribosome for translation.
Since RNA proofreading enzymes are absent, the RNA-dependent RNA transcriptase makes a single nucleotide insertion error roughly every 10 thousand nucleotides, which is the approximate length of the influenza vRNA. Hence, nearly every newly manufactured influenza virus will contain a mutation in its genome.[54] The separation of the genome into eight separate segments of vRNA allows mixing (reassortment) of the genes if more than one variety of influenza virus has infected the same cell (superinfection). The resulting alteration in the genome segments packaged into viral progeny confers new behavior, sometimes the ability to infect new host species or to overcome protective immunity of host populations to its old genome (in which case it is called an antigenic shift).[16]
Viability and disinfection
Mammalian influenza viruses tend to be labile, but can survive several hours in mucus.[55] Avian influenza virus can survive for 100 days in distilled water at room temperature, and 200 days at 17 °C (63 °F). The avian virus is inactivated more quickly in manure, but can survive for up to 2 weeks in feces on cages. Avian influenza viruses can survive indefinitely when frozen.[55] Influenza viruses are susceptible to bleach, 70% ethanol, aldehydes, oxidizing agents, and quaternary ammonium compounds. They are inactivated by heat of 133 °F (56 °C) for minimum of 60 minutes, as well as by low pH <2.[55]
Vaccination and prophylaxis
Vaccines and drugs are available for the prophylaxis and treatment of influenza virus infections. Vaccines are composed of either inactivated or live attenuated virions of the H1N1 and H3N2 human influenza A viruses, as well as those of influenza B viruses. Because the antigenicities of the wild viruses evolve, vaccines are reformulated annually by updating the seed strains. However, when the antigenicities of the seed strains and wild viruses do not match, vaccines fail to protect the vaccinees. In addition, even when they do match, escape mutants are often generated. Drugs available for the treatment of influenza include Amantadine and Rimantadine, which inhibit the uncoating of virions by interfering with M2, and Oseltamivir (marketed under the brand name Tamiflu), Zanamivir, and Peramivir, which inhibit the release of virions from infected cells by interfering with NA. However, escape mutants are often generated for the former drug and less frequently for the latter drug.[56]
See also
- Dog flu
- Influenza-like illness
References
- International Committee on Taxonomy of Viruses Index of Viruses — Orthomyxovirus (2006). In: ICTVdB—The Universal Virus Database, version 4. Büchen-Osmond, C (Ed), Columbia University, New York.
- Jones LD, Nuttall PA (1989). "Non-viraemic transmission of Thogoto virus: influence of time and distance". Trans. R. Soc. Trop. Med. Hyg. 83 (5): 712–14. doi:10.1016/0035-9203(89)90405-7. PMID 2617637.
- Ely B (1999). "Infectious Salmon Anaemia". Mill Hill Essays. National Institute for Medical Research. Archived from the original on 2007-08-24. Retrieved 2007-09-14.
- Raynard RS, Murray AG, Gregory A (2001). "Infectious salmon anaemia virus in wild fish from Scotland". Dis. Aquat. Org. 46 (2): 93–100. doi:10.3354/dao046093. PMID 11678233.
- Biere B, Bauer B, Schweiger B (April 2010). "Differentiation of influenza B virus lineages Yamagata and Victoria by real-time PCR" (PDF). Journal of Clinical Microbiology. 48 (4): 1425–7. doi:10.1128/JCM.02116-09. PMC 2849545. PMID 20107085.
- ICTV Taxonomy History, ICTV, 2014, retrieved 6 June 2006
- ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- Hay A, Gregory V, Douglas A, Lin Y (Dec 29, 2001). "The evolution of human influenza viruses". Philos Trans R Soc Lond B Biol Sci. 356 (1416): 1861–70. doi:10.1098/rstb.2001.0999. PMC 1088562. PMID 11779385.
- "Avian Influenza (Bird Flu)". Centers for Disease Control and Prevention. Retrieved 2007-09-15.
- Kumar, Binod; Asha, Kumari; Khanna, Madhu; Ronsard, Larance; Meseko, Clement Adebajo; Sanicas, Melvin (April 2018). "The emerging influenza virus threat: status and new prospects for its therapy and control". Archives of Virology. 163 (4): 831–844. doi:10.1007/s00705-018-3708-y. ISSN 1432-8798. PMID 29322273.
- Atkinson W, Hamborsky J, McIntyre L, Wolfe S, eds. (2007). Epidemiology and Prevention of Vaccine-Preventable Diseases (10th ed.). Washington DC: Centers for Disease Control and Prevention.
- "Avian Influenza (Bird Flu): Implications for Human Disease". Center for Infectious Disease Research & Policy, University of Minnesota. 2007-06-27. Retrieved 2007-09-14.
- Wang TT, Palese P (June 2009). "Unraveling the Mystery of Swine Influenza Virus". Cell. 137 (6): 983–85. doi:10.1016/j.cell.2009.05.032. PMID 19524497.
- Taubenberger, JK, Morens, DM (April 2009). "Pandemic influenza – including a risk assessment of H5N1". Rev. – Off. Int. Epizoot. 28 (1): 187–202. doi:10.20506/rst.28.1.1879. PMC 2720801. PMID 19618626.
- Fouchier R, Schneeberger P, Rozendaal F, Broekman J, Kemink S, Munster V, Kuiken T, Rimmelzwaan G, Schutten M, Van Doornum G, Koch G, Bosman A, Koopmans M, Osterhaus A (2004). "Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome". Proc Natl Acad Sci USA. 101 (5): 1356–61. Bibcode:2004PNAS..101.1356F. doi:10.1073/pnas.0308352100. PMC 337057. PMID 14745020.
- Hilleman MR (August 2002). "Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control". Vaccine. 20 (25–26): 3068–87. doi:10.1016/S0264-410X(02)00254-2. PMID 12163258.
- Potter CW (October 2001). "A history of influenza". Journal of Applied Microbiology. 91 (4): 572–9. doi:10.1046/j.1365-2672.2001.01492.x. PMID 11576290.
- "Ten things you need to know about pandemic influenza". World Health Organization. 14 October 2005. Archived from the original on 23 September 2009. Retrieved 26 September 2009.
- Valleron AJ, Cori A, Valtat S, Meurisse S, Carrat F, Boëlle PY (May 2010). "Transmissibility and geographic spread of the 1889 influenza pandemic". Proc. Natl. Acad. Sci. USA. 107 (19): 8778–81. Bibcode:2010PNAS..107.8778V. doi:10.1073/pnas.1000886107. PMC 2889325. PMID 20421481.
- Mills CE, Robins JM, Lipsitch M (December 2004). "Transmissibility of 1918 pandemic influenza". Nature. 432 (7019): 904–06. Bibcode:2004Natur.432..904M. doi:10.1038/nature03063. PMID 15602562.
- Donaldson LJ, Rutter PD, Ellis BM, et al. (2009). "Mortality from pandemic A/H1N1 2009 influenza in England: public health surveillance study". BMJ. 339: b5213. doi:10.1136/bmj.b5213. PMC 2791802. PMID 20007665.
- "ECDC Daily Update – Pandemic (H1N1) 2009 – January 18, 2010" (PDF). European Centre for Disease Prevention and Control. 2010-01-18. Archived from the original (PDF) on January 22, 2010. Retrieved 2010-01-18.
- Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, Feikin DR, Fowler KB, Gordon A, Hien NT, Horby P, Huang QS, Katz MA, Krishnan A, Lal R, Montgomery JM, Mølbak K, Pebody R, Presanis AM, Razuri H, Steens A, Tinoco YO, Wallinga J, Yu H, Vong S, Bresee J, Widdowson MA (September 2012). "Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study". The Lancet. Infectious Diseases (Submitted manuscript). 12 (9): 687–95. doi:10.1016/S1473-3099(12)70121-4. PMID 22738893.
- Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA (May 2000). "Influenza B virus in seals". Science. 288 (5468): 1051–3. Bibcode:2000Sci...288.1051O. doi:10.1126/science.288.5468.1051. PMID 10807575.
- Nobusawa E, Sato K (April 2006). "Comparison of the mutation rates of human influenza A and B viruses". Journal of Virology. 80 (7): 3675–8. doi:10.1128/JVI.80.7.3675-3678.2006. PMC 1440390. PMID 16537638.
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (March 1992). "Evolution and ecology of influenza A viruses". Microbiological Reviews. 56 (1): 152–79. PMC 372859. PMID 1579108.
- Zambon MC (November 1999). "Epidemiology and pathogenesis of influenza". The Journal of Antimicrobial Chemotherapy. 44 Suppl B (Suppl B): 3–9. doi:10.1093/jac/44.suppl_2.3. PMID 10877456.
- Matsuzaki Y, Sugawara K, Mizuta K, Tsuchiya E, Muraki Y, Hongo S, Suzuki H, Nakamura K (2002). "Antigenic and genetic characterization of influenza C viruses which caused two outbreaks in Yamagata City, Japan, in 1996 and 1998". J Clin Microbiol. 40 (2): 422–29. doi:10.1128/JCM.40.2.422-429.2002. PMC 153379. PMID 11825952.
- Matsuzaki Y, Katsushima N, Nagai Y, Shoji M, Itagaki T, Sakamoto M, Kitaoka S, Mizuta K, Nishimura H (May 1, 2006). "Clinical features of influenza C virus infection in children". J Infect Dis. 193 (9): 1229–35. doi:10.1086/502973. PMID 16586359.
- Katagiri S, Ohizumi A, Homma M (July 1983). "An outbreak of type C influenza in a children's home". J Infect Dis. 148 (1): 51–56. doi:10.1093/infdis/148.1.51. PMID 6309999.
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F (February 2013). "Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses". PLoS Pathogens. 9 (2): e1003176. doi:10.1371/journal.ppat.1003176. PMC 3567177. PMID 23408893.
- Sheng Z, Ran Z, Wang D, Hoppe AD, Simonson R, Chakravarty S, Hause BM, Li F (February 2014). "Genomic and evolutionary characterization of a novel influenza-C-like virus from swine". Archives of Virology. 159 (2): 249–55. doi:10.1007/s00705-013-1815-3. PMC 5714291. PMID 23942954.
- Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F (January 2015). "Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle". Journal of Virology. 89 (2): 1036–42. doi:10.1128/JVI.02718-14. PMC 4300623. PMID 25355894.
- Noda T (2012-01-03). "Native morphology of influenza virions". Frontiers in Microbiology. 2: 269. doi:10.3389/fmicb.2011.00269. PMC 3249889. PMID 22291683.
- "International Committee on Taxonomy of Viruses (ICTV)".
- Bouvier NM, Palese P (September 2008). "The biology of influenza viruses". Vaccine. 26 Suppl 4: D49–53. doi:10.1016/j.vaccine.2008.07.039. PMC 3074182. PMID 19230160.
- International Committee on Taxonomy of Viruses descriptions of: Orthomyxoviridae Influenzavirus B Influenzavirus C
- Badham MD, Rossman JS (September 2016). "Filamentous Influenza Viruses". Current Clinical Microbiology Reports. 3 (3): 155–161. doi:10.1007/s40588-016-0041-7. PMC 5198887. PMID 28042529.
- Noda T (2012). "Native morphology of influenza virions". Frontiers in Microbiology. 2: 269. doi:10.3389/fmicb.2011.00269. PMC 3249889. PMID 22291683.
- Dadonaite B, Vijayakrishnan S, Fodor E, Bhella D, Hutchinson EC (August 2016). "Filamentous influenza viruses". The Journal of General Virology. 97 (8): 1755–64. doi:10.1099/jgv.0.000535. PMC 5935222. PMID 27365089.
- Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, Bolotov P, Dernovoy D, Tatusova T, Bao Y, St George K, Taylor J, Lipman DJ, Fraser CM, Taubenberger JK, Salzberg SL (October 2005). "Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution". Nature. 437 (7062): 1162–6. Bibcode:2005Natur.437.1162G. doi:10.1038/nature04239. PMID 16208317.
- Suzuki Y (March 2005). "Sialobiology of influenza: molecular mechanism of host range variation of influenza viruses". Biological & Pharmaceutical Bulletin. 28 (3): 399–408. doi:10.1248/bpb.28.399. PMID 15744059.
- Wilson JC, von Itzstein M (July 2003). "Recent strategies in the search for new anti-influenza therapies". Current Drug Targets. 4 (5): 389–408. doi:10.2174/1389450033491019. PMID 12816348.
- Suarez DL, Spackman E, Senne DA, Bulaga L, Welsch AC, Froberg K (2003). "The effect of various disinfectants on detection of avian influenza virus by real time RT-PCR". Avian Diseases. 47 (3 Suppl): 1091–5. doi:10.1637/0005-2086-47.s3.1091. PMID 14575118.
- "Avian Influenza (Bird Flu) Implications for Human Disease. Physical characteristics of influenza A viruses". CIDRAP - Center for Infectious Disease Research and Policy. University of Minnesota.
- "Flu viruses 'can live for decades' on ice". The New Zealand Herald. Reuters. November 30, 2006. Retrieved November 1, 2011.
- Wagner R, Matrosovich M, Klenk H (May–Jun 2002). "Functional balance between haemagglutinin and neuraminidase in influenza virus infections". Rev Med Virol. 12 (3): 159–66. doi:10.1002/rmv.352. PMID 11987141.
- Lakadamyali M, Rust M, Babcock H, Zhuang X (Aug 5, 2003). "Visualizing infection of individual influenza viruses". Proc Natl Acad Sci USA. 100 (16): 9280–85. Bibcode:2003PNAS..100.9280L. doi:10.1073/pnas.0832269100. PMC 170909. PMID 12883000.
- Cros J, Palese P (September 2003). "Trafficking of viral genomic RNA into and out of the nucleus: influenza, Thogoto and Borna disease viruses". Virus Res. 95 (1–2): 3–12. doi:10.1016/S0168-1702(03)00159-X. PMID 12921991.
- Kash J, Goodman A, Korth M, Katze M (July 2006). "Hijacking of the host-cell response and translational control during influenza virus infection". Virus Res. 119 (1): 111–20. doi:10.1016/j.virusres.2005.10.013. PMID 16630668.
- Nayak D, Hui E, Barman S (December 2004). "Assembly and budding of influenza virus". Virus Res. 106 (2): 147–65. doi:10.1016/j.virusres.2004.08.012. PMID 15567494.
- "Cap Snatching". ViralZone. expasy. Retrieved 11 September 2014.
- Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW (April 2009). "The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit". Nature. 458 (7240): 914–8. Bibcode:2009Natur.458..914D. doi:10.1038/nature07745. PMID 19194459.
- Drake J (May 1, 1993). "Rates of spontaneous mutation among RNA viruses". Proc Natl Acad Sci USA. 90 (9): 4171–5. Bibcode:1993PNAS...90.4171D. doi:10.1073/pnas.90.9.4171. PMC 46468. PMID 8387212.
- Spickler AR (February 2016). "Influenza" (PDF). The Center for Food Security and Public Health. Iowa State University. p. 7.
- Suzuki Y (October 2006). "Natural selection on the influenza virus genome". Molecular Biology and Evolution. 23 (10): 1902–11. doi:10.1093/molbev/msl050. PMID 16818477.
Further reading
- Hoyle, L. (1969). The Influenza Viruses. Virology Monographs. 4. Springer-Verlag. ISBN 978-3-211-80892-4. ISSN 0083-6591. OCLC 4053391.
External links
| Wikispecies has information related to Orthomyxoviridae |
- Health-EU Portal EU work to prepare a global response to influenza.
- Influenza Research Database Database of influenza genomic sequences and related information.
- European Commission—Public Health EU coordination on Pandemic (H1N1) 2009
- 3D Influenza-virus-related structures from the EM Data Bank(EMDB)
- Viralzone: Orthomyxoviridae
- ICTV