Causes of autism
Many causes of autism have been proposed, but understanding of the theory of causation of autism and the other autism spectrum disorders (ASD) is incomplete.[1] Research indicates that genetic factors predominate. The heritability of autism, however, is complex, and it is typically unclear which genes are involved.[2] In rare cases, autism is associated with agents that cause birth defects.[3] Many other causes have been proposed, such as childhood immunizations, but numerous epidemiological studies have shown no scientific evidence supporting any link between vaccinations and autism.[4]
Related disorders
Autism involves atypical brain development which often becomes apparent in behavior and social development before a child is three years old. It can be characterized by impairments in social interaction and communication, as well as restricted interests and stereotyped behavior, and the characterization is independent of any underlying neurological defects.[5][6] Other characteristics include repetitive-like tasks seen in behavior and sensory interests.[7] This article uses the terms autism and ASD to denote classical autism and the wider dispersion of symptoms and manifestations of autism, respectively.
Autism's theory of causation is incomplete.[1] It has long been presumed that there is a common cause at the genetic, cognitive, and neural levels for autism's characteristic triad of symptoms.[8] However, there is increasing suspicion among researchers that autism does not have a single cause, but is instead a complex disorder with a set of core aspects that have distinct causes.[8][9] Different underlying brain dysfunctions have been hypothesized to result in the common symptoms of autism, just as completely different brain problems result in intellectual disability. The terms autism or ASDs capture the wide range of disease processes at work.[10] Although these distinct causes have been hypothesized to often co-occur,[9] it has also been suggested that the correlation between the causes has been exaggerated.[11] The number of people known to have autism has increased dramatically since the 1980s, at least partly due to changes in diagnostic practice. It is unknown whether prevalence has increased as well.[12]
The consensus among mainstream autism researchers is that genetic factors predominate. Environmental factors that have been claimed to contribute to autism or exacerbate its symptoms, or that may be important to consider in future research, include certain foods,[13] infectious disease, heavy metals, solvents, diesel exhaust, PCBs, phthalates and phenols used in plastic products, pesticides, brominated flame retardants, alcohol, smoking, and illicit drugs.[12] Among these factors, vaccines have attracted much attention, as parents may first become aware of autistic symptoms in their child around the time of a routine vaccination, and parental concern about vaccines has led to a decreasing uptake of childhood immunizations and an increasing likelihood of measles outbreaks.[14][15] However, there is overwhelming scientific evidence showing no causal association between the measles-mumps-rubella (MMR) vaccine and autism, and there is no scientific evidence that the vaccine preservative thiomersal causes autism.[4][16]
Genetics
Genetic factors may be the most significant cause for autism spectrum disorders. Early studies of twins had estimated heritability to be over 90%, meaning that genetics explains over 90% of whether a child will develop autism.[2] However, this may be an overestimation, as new twin studies estimate the heritability at between 60–90%.[17][18] Many of the non-autistic co-twins had learning or social disabilities. For adult siblings the risk for having one or more features of the broader autism phenotype might be as high as 30%.[19]
However, in spite of the strong heritability, most cases of ASD occur sporadically with no recent evidence of family history. It has been hypothesized that spontaneous de novo mutations in the father's sperm or mother's egg contribute to the likelihood of developing autism.[20] There are two lines of evidence that support this hypothesis. First, individuals with autism have significantly reduced fecundity, they are 20 times less likely to have children than average, thus curtailing the persistence of mutations in ASD genes over multiple generations in a family.[21] Second, the likelihood of having a child develop autism increases with advancing paternal age,[22] and mutations in sperm gradually accumulate throughout a man's life.[23]
The first genes to be definitively shown to contribute to risk for autism were found in the early 1990s by researchers looking at gender-specific forms of autism caused by mutations on the X chromosome. An expansion of the CGG trinucleotide repeat in the promoter of the gene FMR1 in boys causes fragile X syndrome, and at least 20% of boys with this mutation have behaviors consistent with autism spectrum disorder.[24] Mutations that inactivate the gene MECP2 cause Rett syndrome, which is associated with autistic behaviors in girls, and in boys the mutation is embryonic lethal.[25]
Besides these early examples, the role of de novo mutations in ASD first became evident when DNA microarray technologies reached sufficient resolution to allow the detection of copy number variation (CNV) in the human genome.[26][27] CNVs are the most common type of structural variation in the genome, consisting of deletions and duplications of DNA that range in size from a kilobase to a few megabases. Microarray analysis has shown that de novo CNVs occur at a significantly higher rate in sporadic cases of autism as compared to the rate in their typically developing siblings and unrelated controls. A series of studies have shown that gene disrupting de novo CNVs occur approximately four times more frequently in ASD than in controls and contribute to approximately 5–10% of cases.[20][28][29][30] Based on these studies, there are predicted to be 130–234 ASD-related CNV loci.[30] The first whole genome sequencing study to comprehensively catalog de novo structural variation at a much higher resolution than DNA microarray studies has shown that the mutation rate is approximately 20% and not elevated in autism compared to sibling controls.[31] However, structural variants in individuals with autism are much larger and four times more likely to disrupt genes, mirroring findings from CNV studies.[31]
CNV studies were closely followed by exome sequencing studies, which sequence the 1–2% of the genome that codes for proteins (the "exome"). These studies found that de novo gene inactivating mutations were observed in approximately 20% of individuals with autism, compared to 10% of unaffected siblings, suggesting the etiology of ASD is driven by these mutations in around 10% of cases.[32][33][34][35][36][37] There are predicted to be 350-450 genes that significantly increase susceptibility to ASDs when impacted by inactivating de novo mutations.[38] A further 12% of cases are predicted to be caused by protein altering missense mutations that change an amino acid but do not inactivate a gene.[34] Therefore approximately 30% of individuals with autism have a spontaneous de novo large CNV that deletes or duplicates genes, or mutation that changes the amino acid code of an individual gene. A further 5–10% of cases have inherited structural variation at loci known to be associated with autism, and these known structural variants may arise de novo in the parents of affected children.[31]
Tens of genes and CNVs have been definitively identified based on the observation of recurrent mutations in different individuals, and suggestive evidence has been found for over 100 others.[39] The Simons Foundation Autism Research Initiative (SFARI) details the evidence for each genetic locus associated with autism.[40]
These early gene and CNV findings have shown that the cognitive and behavioral features associated with each of the underlying mutations is variable. Each mutation is itself associated with a variety of clinical diagnoses, and can also be found in a small percentage of individuals with no clinical diagnosis.[41][42] Thus the genetic disorders that comprise autism are not autism-specific. The mutations themselves are characterized by considerable variability in clinical outcome and typically only a subset of mutation carriers meet criteria for autism. This variable expressivity results in different individuals with the same mutation varying considerably in the severity of their observed particular trait.[43]
The conclusion of these recent studies of de novo mutation is that the spectrum of autism is breaking up into quanta of individual disorders defined by genetics.[43]
One gene that has been linked to autism is SHANK2.[44] Mutations in this gene act in a dominant fashion. Mutations in this gene appear to cause hyperconnectivity between the neurons.
Epigenetics
Epigenetic mechanisms may increase the risk of autism. Epigenetic changes occur as a result not of DNA sequence changes but of chromosomal histone modification or modification of the DNA bases. Such modifications are known to be affected by environmental factors, including nutrition, drugs, and mental stress.[45] Interest has been expressed in imprinted regions on chromosomes 15q and 7q.[46]
Most data supports a polygenic, epistatic model, meaning that the disorder is caused by two or more genes and that those genes are interacting in a complex manner. Several genes, between two and fifteen in number, have been identified and could potentially contribute to disease susceptibility.[47][48] However, an exact determination of the cause of ASD has yet to be discovered and there probably is not one single genetic cause of any particular set of disorders, leading many researchers to believe that epigenetic mechanisms, such as genomic imprinting or epimutations, may play a major role.[49][50]
Epigenetic mechanisms can contribute to disease phenotypes. Epigenetic modifications include DNA cytosine methylation and post-translational modifications to histones. These mechanisms contribute to regulating gene expression without changing the sequence of the DNA and may be influenced by exposure to environmental factors and may be heritable from parents.[46] Rett syndrome and Fragile X syndrome (FXS) are single gene disorders related to ASD with overlapping symptoms that include deficient neurological development, impaired language and communication, difficulties in social interactions, and stereotyped hand gestures. It is not uncommon for a patient to be diagnosed with both ASD and Rett syndrome and/or FXS. Epigenetic regulatory mechanisms play the central role in pathogenesis of these two diseases.[49][51][52] Rett syndrome is caused by a mutation in the gene that encodes methyl-CpG-binding protein (MECP2), one of the key epigenetic regulators of gene expression.[53] MeCP2 binds methylated cytosine residues in DNA and interacts with complexes that remodel chromatin into repressive structures.[54][55] On the other hand, FXS is caused by mutations that are both genetic and epigenetic. Expansion of the CGG repeat in the 5’-untranslated region of the FMR1 genes leads to susceptibility of epigenetic silencing, leading to loss of gene expression.[52]
Genomic imprinting may also contribute to ASD. Genomic imprinting is another example of epigenetic regulation of gene expression. In this instance, the epigenetic modification(s) causes the offspring to express the maternal copy of a gene or the paternal copy of a gene, but not both. The imprinted gene is silenced through epigenetic mechanisms. Candidate genes and susceptibility alleles for autism are identified using a combination of techniques, including genome-wide and targeted analyses of allele sharing in sib-pairs, using association studies and transmission disequilibrium testing (TDT) of functional and/or positional candidate genes and examination of novel and recurrent cytogenetic aberrations. Results from numerous studies have identified several genomic regions known to be subject to imprinting, candidate genes, and gene-environment interactions. Particularly, chromosomes 15q and 7q appear to be epigenetic hotspots in contributing to ASD. Also, genes on the X chromosome may play an important role, as in Rett Syndrome.[46]
Prenatal environment
The risk of autism is associated with several prenatal risk factors, including advanced age in either parent, diabetes, bleeding, and use of psychiatric drugs in the mother during pregnancy.[56] Autism has been linked to birth defect agents acting during the first eight weeks from conception, though these cases are rare.[57]
Infectious processes
Prenatal viral infection has been called the principal non-genetic cause of autism. Prenatal exposure to rubella or cytomegalovirus activates the mother's immune response and may greatly increase the risk for autism in mice.[58] Congenital rubella syndrome is the most convincing environmental cause of autism.[59] Infection-associated immunological events in early pregnancy may affect neural development more than infections in late pregnancy, not only for autism, but also for psychiatric disorders of presumed neurodevelopmental origin, notably schizophrenia.[60]
Environmental agents
Teratogens are environmental agents that cause birth defects. Some agents that are theorized to cause birth defects have also been suggested as potential autism risk factors, although there is little to no scientific evidence to back such claims. These include exposure of the embryo to valproic acid,[61] paracetamol,[62] thalidomide or misoprostol.[63] These cases are rare.[64] Questions have also been raised whether ethanol (grain alcohol) increases autism risk, as part of fetal alcohol syndrome or alcohol-related birth defects.[63] All known teratogens appear to act during the first eight weeks from conception, and though this does not exclude the possibility that autism can be initiated or affected later, it is strong evidence that autism arises very early in development.[3]
Autoimmune and inflammatory diseases
Maternal inflammatory and autoimmune diseases can damage embryonic and fetal tissues, aggravating a genetic problem or damaging the nervous system.[65]
Other maternal conditions
Thyroid problems that lead to thyroxine deficiency in the mother in weeks 8–12 of pregnancy have been postulated to produce changes in the fetal brain leading to autism. Thyroxine deficiencies can be caused by inadequate iodine in the diet, and by environmental agents that interfere with iodine uptake or act against thyroid hormones. Possible environmental agents include flavonoids in food, tobacco smoke, and most herbicides. This hypothesis has not been tested.[66]
Diabetes in the mother during pregnancy is a significant risk factor for autism; a 2009 meta-analysis found that gestational diabetes was associated with a twofold increased risk. A 2014 review also found that maternal diabetes was significantly associated with an increased risk of ASD.[67] Although diabetes causes metabolic and hormonal abnormalities and oxidative stress, no biological mechanism is known for the association between gestational diabetes and autism risk.[56]
Maternal obesity during pregnancy may also increase the risk of autism, although further study is needed.[68]
Maternal malnutrition during preconception and pregnancy influences fetal neurodevelopment. Intrauterine growth restriction is associated with ASD, in both term and preterm infants.[69]
Other in utero
It has been hypothesized that folic acid taken during pregnancy could play a role in reducing cases of autism by modulating gene expression through an epigenetic mechanism. This hypothesis is supported by multiple studies.[70]
Prenatal stress, consisting of exposure to life events or environmental factors that distress an expectant mother, has been hypothesized to contribute to autism, possibly as part of a gene-environment interaction. Autism has been reported to be associated with prenatal stress both with retrospective studies that examined stressors such as job loss and family discord, and with natural experiments involving prenatal exposure to storms; animal studies have reported that prenatal stress can disrupt brain development and produce behaviors resembling symptoms of autism.[71] However, other studies have cast doubts on this association, notably population based studies in England and Sweden finding no link between stressful life events and ASD.[72]
The fetal testosterone theory hypothesizes that higher levels of testosterone in the amniotic fluid of mothers pushes brain development towards improved ability to see patterns and analyze complex systems while diminishing communication and empathy, emphasizing "male" traits over "female", or in E-S theory terminology, emphasizing "systemizing" over "empathizing". One project has published several reports suggesting that high levels of fetal testosterone could produce behaviors relevant to those seen in autism.[73]
Based in part on animal studies, diagnostic ultrasounds administered during pregnancy have been hypothesized to increase the child's risk of autism. This hypothesis is not supported by independently published research, and examination of children whose mothers received an ultrasound has failed to find evidence of harmful effects.[74]
Some research suggests that maternal exposure to selective serotonin reuptake inhibitors during pregnancy is associated with an increased risk of autism, but it remains unclear whether there is a causal link between the two.[75] There is evidence, for example, that this association may be an artifact of confounding by maternal mental illness.[76]
Perinatal environment
Autism is associated with some perinatal and obstetric conditions. A 2007 review of risk factors found associated obstetric conditions that included low birth weight and gestation duration, and hypoxia during childbirth. This association does not demonstrate a causal relationship. As a result, an underlying cause could explain both autism and these associated conditions.[77] There is growing evidence that perinatal exposure to air pollution may be a risk factor for autism,[78] although this evidence suffers from methodological limitations, including a small number of studies and failure to control for potential confounding factors.[79]
Postnatal environment
A wide variety of postnatal contributors to autism have been proposed, including gastrointestinal or immune system abnormalities, allergies, and exposure of children to drugs, vaccines, infection, certain foods, or heavy metals. The evidence for these risk factors is anecdotal and has not been confirmed by reliable studies.[80]
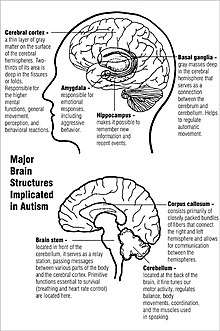
Amygdala neurons
This theory hypothesizes that an early developmental failure involving the amygdala cascades on the development of cortical areas that mediate social perception in the visual domain. The fusiform face area of the ventral stream is implicated. The idea is that it is involved in social knowledge and social cognition, and that the deficits in this network are instrumental in causing autism.[81]
Autoimmune disease
This theory hypothesizes that autoantibodies that target the brain or elements of brain metabolism may cause or exacerbate autism. It is related to the maternal infection theory, except that it postulates that the effect is caused by the individual's own antibodies, possibly due to an environmental trigger after birth. It is also related to several other hypothesized causes; for example, viral infection has been hypothesized to cause autism via an autoimmune mechanism.[82]
Interactions between the immune system and the nervous system begin early during embryogenesis, and successful neurodevelopment depends on a balanced immune response. It is possible that aberrant immune activity during critical periods of neurodevelopment is part of the mechanism of some forms of ASD.[83] A small percentage of autism cases are associated with infection, usually before birth. Results from immune studies have been contradictory. Some abnormalities have been found in specific subgroups, and some of these have been replicated. It is not known whether these abnormalities are relevant to the pathology of autism, for example, by infection or autoimmunity, or whether they are secondary to the disease processes.[84] As autoantibodies are found in diseases other than ASD, and are not always present in ASD,[85] the relationship between immune disturbances and autism remains unclear and controversial.[86] A 2015 systematic review and meta-analysis found that children with a family history of autoimmune diseases were at a greater risk of autism compared to children without such a history.[87]
When an underlying maternal autoimmune disease is present, antibodies circulating to the fetus could contribute to the development of autism spectrum disorders.[88]
Gastrointestinal connection
Gastrointestinal problems are one of the most commonly associated medical disorders in people with autism.[89] These are linked to greater social impairment, irritability, behavior and sleep problems, language impairments and mood changes, so the theory that they are an overlap syndrome has been postulated.[89][90] Studies indicate that gastrointestinal inflammation, immunoglobulin E-mediated or cell-mediated food allergies, gluten-related disorders (celiac disease, wheat allergy, non-celiac gluten sensitivity), visceral hypersensitivity, dysautonomia and gastroesophageal reflux are the mechanisms that possibly link both.[90]
A 2016 review concludes that enteric nervous system abnormalities might play a role in several neurological disorders, including autism. Neural connections and the immune system are a pathway that may allow diseases originated in the intestine to spread to the brain.[91] A 2018 review suggests that the frequent association of gastrointestinal disorders and autism is due to abnormalities of the gut–brain axis.[89]
The "leaky gut" hypothesis is popular among parents of children with autism. It is based on the idea that defects in the intestinal barrier produce an excessive increase of the intestinal permeability, allowing substances present in the intestine, including bacteria, environmental toxins and food antigens, to pass into the blood. The data supporting this theory are limited and contradictory, since both increased intestinal permeability and normal permeability have been documented in people with autism. Studies with mice provide some support to this theory and suggest the importance of intestinal flora, demonstrating that the normalization of the intestinal barrier was associated with an improvement in some of the ASD-like behaviours.[91] Studies on subgroups of people with ASD showed the presence of high plasma levels of zonulin, a protein that regulates permeability opening the "pores" of the intestinal wall, as well as intestinal dysbiosis (reduced levels of Bifidobacteria and increased abundance of Akkermansia muciniphila, Escherichia coli, Clostridia and Candida fungi) that promotes the production of proinflammatory cytokines, all of which produces excessive intestinal permeability.[92] This allows passage of bacterial endotoxins from the gut into the bloodstream, stimulating liver cells to secrete tumor necrosis factor alpha (TNFα), which modulates blood–brain barrier permeability. Studies on ASD people showed that TNFα cascades produce proinflammatory cytokines, leading to peripheral inflammation and activation of microglia in the brain, which indicates neuroinflammation.[92] In addition, neuroactive opioid peptides from digested foods have been shown to leak into the bloodstream and permeate the blood–brain barrier, influencing neural cells and causing autistic symptoms.[92] (See Endogenous opiate precursor theory)
After a preliminary 1998 study of three children with ASD treated with secretin infusion reported improved GI function and dramatic improvement in behavior, many parents sought secretin treatment and a black market for the hormone developed quickly.[93] Later studies found secretin clearly ineffective in treating autism.[94]
Endogenous opiate precursor theory
In 1979, Jaak Panksepp proposed a connection between autism and opiates, noting that injections of minute quantities of opiates in young laboratory animals induce symptoms similar to those observed among autistic children.[95] The possibility of a relationship between autism and the consumption of gluten and casein was first articulated by Kalle Reichelt in 1991.[96]
Opiate theory hypothesizes that autism is the result of a metabolic disorder in which opioid peptides gliadorphin (aka gluteomorphin) and casomorphin, produced through metabolism of gluten (present in wheat and related cereals) and casein (present in dairy products), pass through an abnormally permeable intestinal wall and then proceed to exert an effect on neurotransmission through binding with opioid receptors. It has been postulated that the resulting excess of opioids affects brain maturation, and causes autistic symptoms, including behavioural difficulties, attention problems, and alterations in communicative capacity and social and cognitive functioning.[96][97]
Although high levels of these opioids are eliminated in the urine, it has been suggested that a small part of them cross into the brain causing interference of signal transmission and disruption of normal activity. Three studies have reported that urine samples of people with autism show an increased 24-hour peptide excretion.[96] A study with a control group found no appreciable differences in opioid levels in urine samples of people with autism compared to controls.[92] Two studies showed an increased opioid levels in cerebrospinal fluid of people with autism.[96]
The theory further states that removing opiate precursors from a child's diet may allow time for these behaviors to cease, and neurological development in very young children to resume normally.[98] As of 2014 there is no good evidence that a gluten-free diet is of benefit as a standard treatment for autism.[99][100][101] Problems observed in studies carried out include the suspicion that there were transgressions of the diet because the participants asked for food containing gluten or casein to siblings and peers; and the lack of a washout period, that could diminish the effectiveness of the treatment if gluten or casein peptides have a long term residual effect, which is especially relevant in studies of short duration.[101] In the subset of people who have gluten sensitivity there is limited evidence that suggests that a gluten-free diet may improve some autistic behaviors.[99][102][103]
Lack of vitamin D
The hypothesis that vitamin D deficiency has a role in autism is biologically plausible, but not researched.[104]
Lead
Lead poisoning has been suggested as a possible risk factor for autism, as the lead blood levels of autistic children has been reported to be significantly higher than typical.[105] The atypical eating behaviors of autistic children, along with habitual mouthing and pica, make it hard to determine whether increased lead levels are a cause or a consequence of autism.[105]
Locus coeruleus–noradrenergic system
This theory hypothesizes that autistic behaviors depend at least in part on a developmental dysregulation that results in impaired function of the locus coeruleus–noradrenergic (LC-NA) system. The LC-NA system is heavily involved in arousal and attention; for example, it is related to the brain's acquisition and use of environmental cues.[106]
Mercury
This theory hypothesizes that autism is associated with mercury poisoning, based on perceived similarity of symptoms and reports of mercury or its biomarkers in some autistic children.[107] This view has gained little traction in the scientific community as the typical symptoms of mercury toxicity are significantly different from symptoms seen in autism.[108] The principal source of human exposure to organic mercury is via fish consumption and for inorganic mercury is dental amalgams. The evidence so far is indirect for the association between autism and mercury exposure after birth, as no direct test has been reported, and there is no evidence of an association between autism and postnatal exposure to any neurotoxicant.[109] A meta-analysis published in 2007 concluded that there was no link between mercury and autism.[110]
Oxidative stress
This theory hypothesizes that toxicity and oxidative stress may cause autism in some cases. Evidence includes genetic effects on metabolic pathways, reduced antioxidant capacity, enzyme changes, and enhanced biomarkers for oxidative stress; however, the overall evidence is weaker than it is for involvement oxidative stress with disorders such as schizophrenia.[111] One theory is that stress damages Purkinje cells in the cerebellum after birth, and it is possible that glutathione is involved.[112] Autistic children have lower levels of total glutathione, and higher levels of oxidized glutathione.[113] Based on this theory, antioxidants may be a useful treatment for autism.[114]
Viral infection
Many studies have presented evidence for and against association of autism with viral infection after birth. Laboratory rats infected with Borna disease virus show some symptoms similar to those of autism but blood studies of autistic children show no evidence of infection by this virus. Members of the herpes virus family may have a role in autism, but the evidence so far is anecdotal. Viruses have long been suspected as triggers for immune-mediated diseases such as multiple sclerosis but showing a direct role for viral causation is difficult in those diseases, and mechanisms whereby viral infections could lead to autism are speculative.[58]
Social construct
The social construct theory says that the boundary between normal and abnormal is subjective and arbitrary, so autism does not exist as an objective entity, but only as a social construct. It further argues that autistic individuals themselves have a way of being that is partly socially constructed.[115]
Asperger syndrome and high-functioning autism are particular targets of the theory that social factors determine what it means to be autistic. The theory hypothesizes that individuals with these diagnoses inhabit the identities that have been ascribed to them, and promote their sense of well-being by resisting or appropriating autistic ascriptions.[116]
Discredited theories
Refrigerator mother
Bruno Bettelheim believed that autism was linked to early childhood trauma, and his work was highly influential for decades both in the medical and popular spheres. In his discredited theory, he blamed mothers, of individuals with autism for having caused their child's condition through the withholding of affection.[117] Leo Kanner, who first described autism,[118] suggested that parental coldness might contribute to autism.[119] Although Kanner eventually renounced the theory, Bettelheim put an almost exclusive emphasis on it in both his medical and his popular books. Treatments based on these theories failed to help children with autism, and after Bettelheim's death, it came out that his reported rates of cure (around 85%) were found to be fraudulent.[120]
Vaccines
Scientific studies have refuted a causal relationship between vaccinations and autism.[121][122][123] Despite this, some parents believe that vaccinations cause autism and therefore delay or avoid immunizing their children (for example, under the "vaccine overload" hypothesis that giving many vaccines at once may overwhelm a child's immune system and lead to autism,[124] even though this hypothesis has no scientific evidence and is biologically implausible[125]). Because diseases such as measles can cause severe disabilities and death, the risk of death or disability for an unvaccinated child is higher than the risk for a child who has been vaccinated.[126] Despite all this, antivaccine activism continues. A developing tactic appears to be the "promotion of irrelevant research [as] an active aggregation of several questionable or peripherally related research studies in an attempt to justify the science underlying a questionable claim."[127]
MMR vaccine
The MMR vaccine as a cause of autism is one of the most extensively debated hypotheses regarding the origins of autism. Andrew Wakefield et al. reported a study of 12 children who had autism and bowel symptoms, in some cases reportedly with onset after MMR.[128] Although the paper, which was later retracted by the journal,[128] concluded "We did not prove an association between measles, mumps, and rubella vaccine and the syndrome described,"[129] Wakefield nevertheless suggested during a 1998 press conference that giving children the vaccines in three separate doses would be safer than a single dose.
In 2004, the interpretation of a causal link between MMR vaccine and autism was formally retracted by ten of Wakefield's twelve co-authors.[130] The retraction followed an investigation by The Sunday Times, which stated that Wakefield "acted dishonestly and irresponsibly".[131] The Centers for Disease Control and Prevention,[132] the Institute of Medicine of the National Academy of Sciences,[133] and the U.K. National Health Service[134] have all concluded that there is no evidence of a link between the MMR vaccine and autism.
In February 2010, The Lancet, which published Wakefield's study, fully retracted it after an independent auditor found the study to be flawed.[128] In January 2011, an investigation published in the journal BMJ described the Wakefield study as the result of deliberate fraud and manipulation of data.[135][136][137][138]
Thiomersal (thimerosal)
Perhaps the best-known hypothesis involving mercury and autism involves the use of the mercury-based compound thiomersal, a preservative that has been phased out from most childhood vaccinations in developed countries including US and the EU.[139] Parents may first become aware of autistic symptoms in their child around the time of a routine vaccination. There is no scientific evidence for a causal connection between thiomersal and autism, but parental concern about a relationship between thiomersal and vaccines has led to decreasing rates of childhood immunizations[4] and increasing likelihood of disease outbreaks.[140][141] In 1999, due to concern about the dose of mercury infants were being exposed to, the U.S. Public Health Service recommended that thiomersal be removed from childhood vaccines, and by 2002 the flu vaccine was the only childhood vaccine containing more than trace amounts of thimerosal. Despite this, autism rates did not decrease after the removal of thimerosal, in the US or other countries that also removed thimerosal from their childhood vaccines.[142]
A causal link between thimerosal and autism has been rejected by international scientific and medical professional bodies including the American Medical Association,[143] the American Academy of Pediatrics,[144] the American College of Medical Toxicology,[145] the Canadian Paediatric Society,[146] the U.S. National Academy of Sciences,[133] the Food and Drug Administration,[147] Centers for Disease Control and Prevention,[132] the World Health Organization,[148] the Public Health Agency of Canada,[149] and the European Medicines Agency.[150]
See also
- List of topics characterized as pseudoscience
References
- Trottier G, Srivastava L, Walker CD. Etiology of infantile autism: a review of recent advances in genetic and neurobiological research. J Psychiatry Neurosci. 1999;24(2):103–115. PMID 10212552.
- Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12(1):2–22. doi:10.1038/sj.mp.4001896. PMID 17033636.
- Arndt TL, Stodgell CJ, Rodier PM. The teratology of autism. Int J Dev Neurosci. 2005;23(2–3):189–99. doi:10.1016/j.ijdevneu.2004.11.001. PMID 15749245.
- Doja A, Roberts W. Immunizations and autism: a review of the literature. Can J Neurol Sci. 2006;33(4):341–346. doi:10.1017/s031716710000528x. PMID 17168158.
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th, text revision (DSM-IV-TR) ed. 2000 [Retrieved 2009-02-17]. ISBN 0-89042-025-4. Diagnostic criteria for 299.00 Autistic Disorder.
- World Health Organization. International Statistical Classification of Diseases and Related Health Problems. 10th (ICD-10) ed. 2006 [Retrieved 2007-06-25]. F84. Pervasive developmental disorders.
- McPartland, James C.; Law, Karen; Dawson, Geraldine (August 26, 2015). Autism Spectrum Disorder. Encyclopedia of Mental Health (Second Edition). pp. 124–130. doi:10.1016/B978-0-12-397045-9.00230-5. ISBN 9780123977533.
- Happé F, Ronald A. The 'fractionable autism triad': a review of evidence from behavioural, genetic, cognitive and neural research. Neuropsychol Rev. 2008;18(4):287–304. doi:10.1007/s11065-008-9076-8. PMID 18956240.
- Happé F, Ronald A, Plomin R. Time to give up on a single explanation for autism. Nat Neurosci. 2006;9(10):1218–1220. doi:10.1038/nn1770. PMID 17001340.
- Geschwind DH. Advances in autism. Annu Rev Med. 2009;60:367–380. doi:10.1146/annurev.med.60.053107.121225. PMID 19630577.
- Mandy WP, Skuse DH. What is the association between the social-communication element of autism and repetitive interests, behaviours and activities? J Child Psychol Psychiatry. 2008;49(8):795–808. doi:10.1111/j.1469-7610.2008.01911.x. PMID 18564070.
- Newschaffer CJ, Croen LA, Daniels J et al.. The epidemiology of autism spectrum disorders [PDF]. Annu Rev Public Health. 2007 [Retrieved 2009-10-10];28:235–258. doi:10.1146/annurev.publhealth.28.021406.144007. PMID 17367287.
- Christison GW, Ivany K. Elimination diets in autism spectrum disorders: any wheat amidst the chaff? J Dev Behav Pediatr. 2006;27(2 Suppl 2):S162–171. doi:10.1097/00004703-200604002-00015. PMID 16685183.
- https://www.livescience.com/64728-measles-outbreak-spurs-vaccination.html
- https://globalnews.ca/news/4948647/measles-vaccinations-spike-outbreak-anti-vaxxer-hotspot/
- Schultz ST. Does thimerosal or other mercury exposure increase the risk for autism? A review of current literature.. Acta Neurobiologiae Experimentalis. 2010;70(2):187–195. PMID 20628442.
- Hallmayer, Joachim (1 November 2011). "Genetic Heritability and Shared Environmental Factors Among Twin Pairs With Autism". Archives of General Psychiatry. 68 (11): 1095–1102. doi:10.1001/archgenpsychiatry.2011.76. PMC 4440679. PMID 21727249.
- Ronald, Angelica; Hoekstra, Rosa A. (April 2011). "Autism spectrum disorders and autistic traits: A decade of new twin studies". American Journal of Medical Genetics Part B. 156 (3): 255–274. doi:10.1002/ajmg.b.31159. PMID 21438136.
- Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2(12):943–955. doi:10.1038/35103559. PMID 11733747.
- Sebat, J.; Lakshmi, B.; Malhotra, D.; Troge, J.; Lese-Martin, C.; Walsh, T.; Yamrom, B.; Yoon, S.; Krasnitz, A.; Kendall, J.; Leotta, A.; Pai, D.; Zhang, R.; Lee, Y.-H.; Hicks, J.; Spence, S. J.; Lee, A. T.; Puura, K.; Lehtimaki, T.; Ledbetter, D.; Gregersen, P. K.; Bregman, J.; Sutcliffe, J. S.; Jobanputra, V.; Chung, W.; Warburton, D.; King, M.-C.; Skuse, D.; Geschwind, D. H.; Gilliam, T. C.; Ye, K.; Wigler, M. (20 April 2007). "Strong Association of De Novo Copy Number Mutations with Autism". Science. 316 (5823): 445–449. doi:10.1126/science.1138659. PMC 2993504. PMID 17363630.
- Uher, R (25 August 2009). "The role of genetic variation in the causation of mental illness: an evolution-informed framework". Molecular Psychiatry. 14 (12): 1072–1082. doi:10.1038/mp.2009.85. PMID 19704409.
- Hultman, C M; Sandin, S; Levine, S Z; Lichtenstein, P; Reichenberg, A (30 November 2010). "Advancing paternal age and risk of autism: new evidence from a population-based study and a meta-analysis of epidemiological studies". Molecular Psychiatry. 16 (12): 1203–1212. doi:10.1038/mp.2010.121. PMID 21116277.
- Kong, Augustine; Frigge, Michael L.; Masson, Gisli; Besenbacher, Soren; Sulem, Patrick; Magnusson, Gisli; Gudjonsson, Sigurjon A.; Sigurdsson, Asgeir; Jonasdottir, Aslaug; Jonasdottir, Adalbjorg; Wong, Wendy S. W.; Sigurdsson, Gunnar; Walters, G. Bragi; Steinberg, Stacy; Helgason, Hannes; Thorleifsson, Gudmar; Gudbjartsson, Daniel F.; Helgason, Agnar; Magnusson, Olafur Th.; Thorsteinsdottir, Unnur; Stefansson, Kari (22 August 2012). "Rate of de novo mutations and the importance of father's age to disease risk". Nature. 488 (7412): 471–475. doi:10.1038/nature11396. PMC 3548427. PMID 22914163.
- Hatton, Deborah D.; Sideris, John; Skinner, Martie; Mankowski, Jean; Bailey, Donald B.; Roberts, Jane; Mirrett, Penny (1 September 2006). "Autistic behavior in children with fragile X syndrome: Prevalence, stability, and the impact of FMRP". American Journal of Medical Genetics Part A. 140A (17): 1804–1813. doi:10.1002/ajmg.a.31286. PMID 16700053.
- Zoghbi, Huda Y.; Amir, Ruthie E.; Van den Veyver, Ignatia B.; Wan, Mimi; Tran, Charles Q.; Francke, Uta (1 October 1999). "Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2". Nature Genetics. 23 (2): 185–188. doi:10.1038/13810. PMID 10508514.
- Sebat, J. (23 July 2004). "Large-Scale Copy Number Polymorphism in the Human Genome". Science. 305 (5683): 525–528. doi:10.1126/science.1098918. PMID 15273396.
- Iafrate, A John; Feuk, Lars; Rivera, Miguel N; Listewnik, Marc L; Donahoe, Patricia K; Qi, Ying; Scherer, Stephen W; Lee, Charles (1 August 2004). "Detection of large-scale variation in the human genome". Nature Genetics. 36 (9): 949–951. doi:10.1038/ng1416. PMID 15286789.
- Pinto, Dalila; Delaby, Elsa; Merico, Daniele; Barbosa, Mafalda; Merikangas, Alison; Klei, Lambertus; Thiruvahindrapuram, Bhooma; Xu, Xiao; Ziman, Robert; Wang, Zhuozhi; Vorstman, Jacob A.S.; Thompson, Ann; Regan, Regina; Pilorge, Marion; Pellecchia, Giovanna; Pagnamenta, Alistair T.; Oliveira, Bárbara; Marshall, Christian R.; Magalhaes, Tiago R.; Lowe, Jennifer K.; Howe, Jennifer L.; Griswold, Anthony J.; Gilbert, John; Duketis, Eftichia; Dombroski, Beth A.; De Jonge, Maretha V.; Cuccaro, Michael; Crawford, Emily L.; Correia, Catarina T.; et al. (May 2014). "Convergence of Genes and Cellular Pathways Dysregulated in Autism Spectrum Disorders". The American Journal of Human Genetics. 94 (5): 677–694. doi:10.1016/j.ajhg.2014.03.018. PMC 4067558. PMID 24768552.
- Levy, Dan; Ronemus, Michael; Yamrom, Boris; Lee, Yoon-ha; Leotta, Anthony; Kendall, Jude; Marks, Steven; Lakshmi, B.; Pai, Deepa; Ye, Kenny; Buja, Andreas; Krieger, Abba; Yoon, Seungtai; Troge, Jennifer; Rodgers, Linda; Iossifov, Ivan; Wigler, Michael (June 2011). "Rare De Novo and Transmitted Copy-Number Variation in Autistic Spectrum Disorders". Neuron. 70 (5): 886–897. doi:10.1016/j.neuron.2011.05.015. PMID 21658582.
- Sanders, Stephan J.; Ercan-Sencicek, A. Gulhan; Hus, Vanessa; Luo, Rui; Murtha, Michael T.; Moreno-De-Luca, Daniel; Chu, Su H.; Moreau, Michael P.; Gupta, Abha R.; Thomson, Susanne A.; Mason, Christopher E.; Bilguvar, Kaya; Celestino-Soper, Patricia B.S.; Choi, Murim; Crawford, Emily L.; Davis, Lea; Davis Wright, Nicole R.; Dhodapkar, Rahul M.; DiCola, Michael; DiLullo, Nicholas M.; Fernandez, Thomas V.; Fielding-Singh, Vikram; Fishman, Daniel O.; Frahm, Stephanie; Garagaloyan, Rouben; Goh, Gerald S.; Kammela, Sindhuja; Klei, Lambertus; Lowe, Jennifer K.; Lund, Sabata C.; McGrew, Anna D.; Meyer, Kyle A.; Moffat, William J.; Murdoch, John D.; O'Roak, Brian J.; Ober, Gordon T.; Pottenger, Rebecca S.; Raubeson, Melanie J.; Song, Youeun; Wang, Qi; Yaspan, Brian L.; Yu, Timothy W.; Yurkiewicz, Ilana R.; Beaudet, Arthur L.; Cantor, Rita M.; Curland, Martin; Grice, Dorothy E.; Günel, Murat; Lifton, Richard P.; Mane, Shrikant M.; Martin, Donna M.; Shaw, Chad A.; Sheldon, Michael; Tischfield, Jay A.; Walsh, Christopher A.; Morrow, Eric M.; Ledbetter, David H.; Fombonne, Eric; Lord, Catherine; Martin, Christa Lese; Brooks, Andrew I.; Sutcliffe, James S.; Cook, Edwin H.; Geschwind, Daniel; Roeder, Kathryn; Devlin, Bernie; State, Matthew W. (June 2011). "Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism". Neuron. 70 (5): 863–885. doi:10.1016/j.neuron.2011.05.002. PMC 3939065. PMID 21658581.
- Brandler, William M.; Antaki, Danny; Gujral, Madhusudan; Noor, Amina; Rosanio, Gabriel; Chapman, Timothy R.; Barrera, Daniel J.; Lin, Guan Ning; Malhotra, Dheeraj; Watts, Amanda C.; Wong, Lawrence C.; Estabillo, Jasper A.; Gadomski, Therese E.; Hong, Oanh; Fajardo, Karin V. Fuentes; Bhandari, Abhishek; Owen, Renius; Baughn, Michael; Yuan, Jeffrey; Solomon, Terry; Moyzis, Alexandra G.; Maile, Michelle S.; Sanders, Stephan J.; Reiner, Gail E.; Vaux, Keith K.; Strom, Charles M.; Zhang, Kang; Muotri, Alysson R.; Akshoomoff, Natacha; Leal, Suzanne M.; Pierce, Karen; Courchesne, Eric; Iakoucheva, Lilia M.; Corsello, Christina; Sebat, Jonathan (March 2016). "Frequency and Complexity of De Novo Structural Mutation in Autism". The American Journal of Human Genetics. 98 (4): 667–679. doi:10.1016/j.ajhg.2016.02.018. PMC 4833290. PMID 27018473.
- Iossifov, Ivan; Ronemus, Michael; Levy, Dan; Wang, Zihua; Hakker, Inessa; Rosenbaum, Julie; Yamrom, Boris; Lee, Yoon-ha; Narzisi, Giuseppe; Leotta, Anthony; Kendall, Jude; Grabowska, Ewa; Ma, Beicong; Marks, Steven; Rodgers, Linda; Stepansky, Asya; Troge, Jennifer; Andrews, Peter; Bekritsky, Mitchell; Pradhan, Kith; Ghiban, Elena; Kramer, Melissa; Parla, Jennifer; Demeter, Ryan; Fulton, Lucinda L.; Fulton, Robert S.; Magrini, Vincent J.; Ye, Kenny; Darnell, Jennifer C.; Darnell, Robert B.; Mardis, Elaine R.; Wilson, Richard K.; Schatz, Michael C.; McCombie, W. Richard; Wigler, Michael (April 2012). "De Novo Gene Disruptions in Children on the Autistic Spectrum". Neuron. 74 (2): 285–299. doi:10.1016/j.neuron.2012.04.009. PMC 3619976. PMID 22542183.
- De Rubeis, Silvia; He, Xin; Goldberg, Arthur P.; Poultney, Christopher S.; Samocha, Kaitlin; Ercument Cicek, A.; Kou, Yan; Liu, Li; Fromer, Menachem; Walker, Susan; Singh, Tarjinder; Klei, Lambertus; Kosmicki, Jack; Fu, Shih-Chen; Aleksic, Branko; Biscaldi, Monica; Bolton, Patrick F.; Brownfeld, Jessica M.; Cai, Jinlu; Campbell, Nicholas G.; Carracedo, Angel; Chahrour, Maria H.; Chiocchetti, Andreas G.; Coon, Hilary; Crawford, Emily L.; Crooks, Lucy; Curran, Sarah R.; Dawson, Geraldine; Duketis, Eftichia; Fernandez, Bridget A.; Gallagher, Louise; Geller, Evan; Guter, Stephen J.; Sean Hill, R.; Ionita-Laza, Iuliana; Jimenez Gonzalez, Patricia; Kilpinen, Helena; Klauck, Sabine M.; Kolevzon, Alexander; Lee, Irene; Lei, Jing; Lehtimäki, Terho; Lin, Chiao-Feng; Ma’ayan, Avi; Marshall, Christian R.; McInnes, Alison L.; Neale, Benjamin; Owen, Michael J.; Ozaki, Norio; Parellada, Mara; Parr, Jeremy R.; Purcell, Shaun; Puura, Kaija; Rajagopalan, Deepthi; Rehnström, Karola; Reichenberg, Abraham; Sabo, Aniko; Sachse, Michael; Sanders, Stephan J.; Schafer, Chad; Schulte-Rüther, Martin; Skuse, David; Stevens, Christine; Szatmari, Peter; Tammimies, Kristiina; Valladares, Otto; Voran, Annette; Wang, Li-San; Weiss, Lauren A.; Jeremy Willsey, A.; Yu, Timothy W.; Yuen, Ryan K. C.; Cook, Edwin H.; Freitag, Christine M.; Gill, Michael; Hultman, Christina M.; Lehner, Thomas; Palotie, Aarno; Schellenberg, Gerard D.; Sklar, Pamela; State, Matthew W.; Sutcliffe, James S.; Walsh, Christopher A.; Scherer, Stephen W.; Zwick, Michael E.; Barrett, Jeffrey C.; Cutler, David J.; Roeder, Kathryn; Devlin, Bernie; Daly, Mark J.; Buxbaum, Joseph D. (29 October 2014). "Synaptic, transcriptional and chromatin genes disrupted in autism". Nature. 515 (7526): 209–215. doi:10.1038/nature13772. PMC 4402723. PMID 25363760.
- Iossifov, Ivan; O’Roak, Brian J.; Sanders, Stephan J.; Ronemus, Michael; Krumm, Niklas; Levy, Dan; Stessman, Holly A.; Witherspoon, Kali T.; Vives, Laura; Patterson, Karynne E.; Smith, Joshua D.; Paeper, Bryan; Nickerson, Deborah A.; Dea, Jeanselle; Dong, Shan; Gonzalez, Luis E.; Mandell, Jeffrey D.; Mane, Shrikant M.; Murtha, Michael T.; Sullivan, Catherine A.; Walker, Michael F.; Waqar, Zainulabedin; Wei, Liping; Willsey, A. Jeremy; Yamrom, Boris; Lee, Yoon-ha; Grabowska, Ewa; Dalkic, Ertugrul; Wang, Zihua; Marks, Steven; Andrews, Peter; Leotta, Anthony; Kendall, Jude; Hakker, Inessa; Rosenbaum, Julie; Ma, Beicong; Rodgers, Linda; Troge, Jennifer; Narzisi, Giuseppe; Yoon, Seungtai; Schatz, Michael C.; Ye, Kenny; McCombie, W. Richard; Shendure, Jay; Eichler, Evan E.; State, Matthew W.; Wigler, Michael (29 October 2014). "The contribution of de novo coding mutations to autism spectrum disorder". Nature. 515 (7526): 216–221. doi:10.1038/nature13908. PMC 4313871. PMID 25363768.
- Neale, Benjamin M.; Kou, Yan; Liu, Li; Ma’ayan, Avi; Samocha, Kaitlin E.; Sabo, Aniko; Lin, Chiao-Feng; Stevens, Christine; Wang, Li-San; Makarov, Vladimir; Polak, Paz; Yoon, Seungtai; Maguire, Jared; Crawford, Emily L.; Campbell, Nicholas G.; Geller, Evan T.; Valladares, Otto; Schafer, Chad; Liu, Han; Zhao, Tuo; Cai, Guiqing; Lihm, Jayon; Dannenfelser, Ruth; Jabado, Omar; Peralta, Zuleyma; Nagaswamy, Uma; Muzny, Donna; Reid, Jeffrey G.; Newsham, Irene; Wu, Yuanqing; Lewis, Lora; Han, Yi; Voight, Benjamin F.; Lim, Elaine; Rossin, Elizabeth; Kirby, Andrew; Flannick, Jason; Fromer, Menachem; Shakir, Khalid; Fennell, Tim; Garimella, Kiran; Banks, Eric; Poplin, Ryan; Gabriel, Stacey; DePristo, Mark; Wimbish, Jack R.; Boone, Braden E.; Levy, Shawn E.; Betancur, Catalina; Sunyaev, Shamil; Boerwinkle, Eric; Buxbaum, Joseph D.; Cook Jr, Edwin H.; Devlin, Bernie; Gibbs, Richard A.; Roeder, Kathryn; Schellenberg, Gerard D.; Sutcliffe, James S.; Daly, Mark J. (4 April 2012). "Patterns and rates of exonic de novo mutations in autism spectrum disorders". Nature. 485 (7397): 242–245. doi:10.1038/nature11011. PMC 3613847. PMID 22495311.
- Sanders, Stephan J.; Murtha, Michael T.; Gupta, Abha R.; Murdoch, John D.; Raubeson, Melanie J.; Willsey, A. Jeremy; Ercan-Sencicek, A. Gulhan; DiLullo, Nicholas M.; Parikshak, Neelroop N.; Stein, Jason L.; Walker, Michael F.; Ober, Gordon T.; Teran, Nicole A.; Song, Youeun; El-Fishawy, Paul; Murtha, Ryan C.; Choi, Murim; Overton, John D.; Bjornson, Robert D.; Carriero, Nicholas J.; Meyer, Kyle A.; Bilguvar, Kaya; Mane, Shrikant M.; Šestan, Nenad; Lifton, Richard P.; Günel, Murat; Roeder, Kathryn; Geschwind, Daniel H.; Devlin, Bernie; State, Matthew W. (4 April 2012). "De novo mutations revealed by whole-exome sequencing are strongly associated with autism". Nature. 485 (7397): 237–241. doi:10.1038/nature10945. PMC 3667984. PMID 22495306.
- O’Roak, Brian J.; Vives, Laura; Girirajan, Santhosh; Karakoc, Emre; Krumm, Niklas; Coe, Bradley P.; Levy, Roie; Ko, Arthur; Lee, Choli; Smith, Joshua D.; Turner, Emily H.; Stanaway, Ian B.; Vernot, Benjamin; Malig, Maika; Baker, Carl; Reilly, Beau; Akey, Joshua M.; Borenstein, Elhanan; Rieder, Mark J.; Nickerson, Deborah A.; Bernier, Raphael; Shendure, Jay; Eichler, Evan E. (4 April 2012). "Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations". Nature. 485 (7397): 246–250. doi:10.1038/nature10989. PMC 3350576. PMID 22495309.
- Ronemus, Michael; Iossifov, Ivan; Levy, Dan; Wigler, Michael (16 January 2014). "The role of de novo mutations in the genetics of autism spectrum disorders". Nature Reviews Genetics. 15 (2): 133–141. doi:10.1038/nrg3585. PMID 24430941.
- Betancur, Catalina (March 2011). "Etiological heterogeneity in autism spectrum disorders: More than 100 genetic and genomic disorders and still counting". Brain Research. 1380: 42–77. doi:10.1016/j.brainres.2010.11.078. PMID 21129364.
- "SFARI Gene". SFARI gene. Archived from the original on 2016-04-01. Retrieved 2016-04-13.
- Stefansson, Hreinn; Meyer-Lindenberg, Andreas; Steinberg, Stacy; Magnusdottir, Brynja; Morgen, Katrin; Arnarsdottir, Sunna; Bjornsdottir, Gyda; Walters, G. Bragi; Jonsdottir, Gudrun A.; Doyle, Orla M.; Tost, Heike; Grimm, Oliver; Kristjansdottir, Solveig; Snorrason, Heimir; Davidsdottir, Solveig R.; Gudmundsson, Larus J.; Jonsson, Gudbjorn F.; Stefansdottir, Berglind; Helgadottir, Isafold; Haraldsson, Magnus; Jonsdottir, Birna; Thygesen, Johan H.; Schwarz, Adam J.; Didriksen, Michael; Stensbøl, Tine B.; Brammer, Michael; Kapur, Shitij; Halldorsson, Jonas G.; Hreidarsson, Stefan; Saemundsen, Evald; Sigurdsson, Engilbert; Stefansson, Kari (18 December 2013). "CNVs conferring risk of autism or schizophrenia affect cognition in controls". Nature. 505 (7483): 361–366. doi:10.1038/nature12818. PMID 24352232.
- Shinawi, M.; Liu, P.; Kang, S. H. L.; Shen, J.; Belmont, J. W.; Scott, D. A.; Probst, F. J.; Craigen, W. J.; Graham, B. H.; Pursley, A.; Clark, G.; Lee, J.; Proud, M.; Stocco, A.; Rodriguez, D. L.; Kozel, B. A.; Sparagana, S.; Roeder, E. R.; McGrew, S. G.; Kurczynski, T. W.; Allison, L. J.; Amato, S.; Savage, S.; Patel, A.; Stankiewicz, P.; Beaudet, A. L.; Cheung, S. W.; Lupski, J. R. (12 November 2009). "Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size". Journal of Medical Genetics. 47 (5): 332–341. doi:10.1136/jmg.2009.073015. PMC 3158566. PMID 19914906.
- Brandler, William M.; Sebat, Jonathan (14 January 2015). "From De Novo Mutations to Personalized Therapeutic Interventions in Autism". Annual Review of Medicine. 66 (1): 487–507. doi:10.1146/annurev-med-091113-024550. PMID 25587659.
- Zaslavsky K, Zhang WB, McCready FP, Rodrigues DC, Deneault E, Loo C, Zhao M, Ross PJ, El Hajjar J, Romm A, Thompson T, Piekna A, Wei W, Wang Z, Khattak S, Mufteev M, Pasceri P, Scherer SW, Salter MW, Ellis J (2019) SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons. Nat Neurosci 22(4):556-564
- Miyake K, Hirasawa T, Koide T, Kubota T. Epigenetics in autism and other neurodevelopmental diseases. Adv. Exp. Med. Biol.. 2012;724:91–8. doi:10.1007/978-1-4614-0653-2_7. PMID 22411236.
- Schanen NC. Epigenetics of autism spectrum disorders. Hum. Mol. Genet.. October 2006;15 Spec No 2:R138–50. doi:10.1093/hmg/ddl213. PMID 16987877.
- Pickles, A.; Bolton, P.; Macdonald, H.; Bailey, A.; Le Couteur, A.; Sim, C.H. & Rutter, M. (1995). "Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism". American Journal of Human Genetics. 57 (3): 717–726. PMC 1801262. PMID 7668301.
- Risch N; Spiker D; Lotspeich L; et al. (August 1999). "A genomic screen of autism: evidence for a multilocus etiology". American Journal of Human Genetics. 65 (2): 493–507. doi:10.1086/302497. PMC 1377948. PMID 10417292.
- Samaco, R.C.; Hogart, A. & LaSalle, J.M. (2005). "Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3". Human Molecular Genetics. 14 (4): 483–492. doi:10.1093/hmg/ddi045. PMC 1224722. PMID 15615769.
- Jiang YH; Sahoo T; Michaelis RC; Bercovich D; Bressler J; Kashork CD; Liu Q; Shaffer LG; Schroer RJ; Stockton DW; Spielman RS; Stevenson RE; Beaudet AL (2004). "A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A". American Journal of Medical Genetics. 131 (1): 1–10. doi:10.1002/ajmg.a.30297. PMID 15389703.
- Lopez-Rangel, E. & Lewis, M.E. (2006). "Further evidence for pigenetic influence of MECP2 in Rett, autism and Angelman's syndromes". Clinical Genetics. 69: 23–25. doi:10.1111/j.1399-0004.2006.00543c.x.
- Hagerman, R.J.; Ono, M.Y. & Hagerman, P.J. (2005). "Recent advances in fragile X: a model for autism and neurodegeneration". Current Opinion in Psychiatry. 18 (5): 490–496. doi:10.1097/01.yco.0000179485.39520.b0. PMID 16639106.
- Amir, R.E.; Van den Veyver, I.B.; Wan, M.; Tran, C.Q.; Francke, U. & Zoghbi, H.Y. (October 1999). "Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2". Nature Genetics. 23 (2): 185–188. doi:10.1038/13810. PMID 10508514.
- Klose, R.J. & Bird, A.P. (2006). "Genomic DNA methylation: the mark and its mediators". Trends in Biochemical Sciences. 31 (2): 89–97. doi:10.1016/j.tibs.2005.12.008. PMID 16403636.
- Kriaucionis, S. & Bird, A. (2003). "DNA methylation and Rett syndrome". Human Molecular Genetics. 12 (2): R221–R227. doi:10.1093/hmg/ddg286. PMID 12928486.
- Gardener H, Spiegelman D, Buka SL. Prenatal risk factors for autism: comprehensive meta-analysis. Br J Psychiatry. 2009;195(1):7–14. doi:10.1192/bjp.bp.108.051672. PMID 19567888.
- Roullet FI, Lai JK, Foster JA. In utero exposure to valproic acid and autism--a current review of clinical and animal studies. Neurotoxicol Teratol. 2013;36:47–56. doi:10.1016/j.ntt.2013.01.004. PMID 23395807.
- Libbey JE, Sweeten TL, McMahon WM, Fujinami RS. Autistic disorder and viral infections. J Neurovirol. 2005;11(1):1–10. doi:10.1080/13550280590900553. PMID 15804954.
- Mendelsohn NJ, Schaefer GB. Genetic evaluation of autism. Semin Pediatr Neurol. 2008;15(1):27–31. doi:10.1016/j.spen.2008.01.005. PMID 18342258.
- Meyer U, Yee BK, Feldon J. The neurodevelopmental impact of prenatal infections at different times of pregnancy: the earlier the worse?. Neuroscientist. 2007;13(3):241–56. doi:10.1177/1073858406296401. PMID 17519367.
- Chomiak T, Turner N, Hu B. What We Have Learned about Autism Spectrum Disorder from Valproic Acid. Pathol Res Int. 2013;2013:712758. doi:10.1155/2013/712758. PMID 24381784.
- Avella-Garcia CB, Julvez J, Fortuny J, Rebordosa C, García-Esteban R, Galán IR, Tardón A, Rodríguez-Bernal CL, Iñiguez C, Andiarena A, Santa-Marina L, Sunyer J. Acetaminophen use in pregnancy and neurodevelopment: attention function and autism spectrum symptoms. Int J Epidemiol. doi:10.1093/ije/dyw115. PMID 27353198.
- Dufour-Rainfray D, Vourc'h P, Tourlet S, Guilloteau D, Chalon S, Andres CR. Fetal exposure to teratogens: evidence of genes involved in autism. Neurosci Biobehav Rev. 2011;35(5):1254–65. doi:10.1016/j.neubiorev.2010.12.013. PMID 21195109.
- Miller MT, Strömland K, Ventura L, Johansson M, Bandim JM, Gillberg C. Autism associated with conditions characterized by developmental errors in early embryogenesis: a mini review. Int. J. Dev. Neurosci.. 2005;23(2-3):201–19. doi:10.1016/j.ijdevneu.2004.06.007. PMID 15749246.
- Samsam M, Ahangari R, Naser SA (2014). "Pathophysiology of autism spectrum disorders: revisiting gastrointestinal involvement and immune imbalance". World J Gastroenterol (Review). 20 (29): 9942–51. doi:10.3748/wjg.v20.i29.9942. PMC 4123375. PMID 25110424.
- Román GC. Autism: transient in utero hypothyroxinemia related to maternal flavonoid ingestion during pregnancy and to other environmental antithyroid agents. J Neurol Sci. 2007;262(1–2):15–26. doi:10.1016/j.jns.2007.06.023. PMID 17651757.
- Xu, Guifeng. Maternal Diabetes and the Risk of Autism Spectrum Disorders in the Offspring: A Systematic Review and Meta-Analysis. Journal of Autism and Developmental Disorders. 22 September 2013;44(4):766–775. doi:10.1007/s10803-013-1928-2. PMID 24057131.
- Li YM et al.. Association Between Maternal Obesity and Autism Spectrum Disorder in Offspring: A Meta-analysis. J Autism Dev Disord. 2015. doi:10.1007/s10803-015-2549-8. PMID 26254893.
- Vohr BR, Poggi Davis E, Wanke CA, Krebs NF (2017). "Neurodevelopment: The Impact of Nutrition and Inflammation During Preconception and Pregnancy in Low-Resource Settings". Pediatrics (Review). 139 (Suppl 1): S38–S49. doi:10.1542/peds.2016-2828F. PMID 28562247.
- Lyall K, Schimdt RJ, Hertz-Picciotto I. Maternal lifestyle and environmental risk factors for autism spectrum disorders. International Journal of Epidemiology. 11 February 2014;43(2):443–464. doi:10.1093/ije/dyt282. PMID 24518932.
- Kinney DK, Munir KM, Crowley DJ, Miller AM. Prenatal stress and risk for autism. Neurosci Biobehav Rev. 2008;32(8):1519–32. doi:10.1016/j.neubiorev.2008.06.004. PMID 18598714.
- Prenatal and Early Life Exposure to Stressful Life Events and Risk of Autism Spectrum Disorders: Population-Based Studies in Sweden and England. PLOS ONE. 2012;7(6):e38893. doi:10.1371/journal.pone.0038893. PMID 22719977.
- Fetal testosterone and autistic traits:
- Auyeung B, Baron-Cohen S. A role for fetal testosterone in human sex differences. In: Zimmerman AW. Autism: Current Theories and Evidence. Humana; 2009. doi:10.1007/978-1-60327-489-0_8. ISBN 978-1-60327-488-3. p. 185–208.
- Manson JE. Prenatal exposure to sex steroid hormones and behavioral/cognitive outcomes. Metabolism. 2008;57(Suppl 2):S16–21. doi:10.1016/j.metabol.2008.07.010. PMID 18803959.
- Abramowicz JS. Ultrasound and autism: association, link, or coincidence?. J Ultrasound Med. 2012;31(8):1261–9. PMID 22837291.
- Man KK, Tong HH, Wong LY, Chan EW, Simonoff E, Wong IC. Exposure to selective serotonin reuptake inhibitors during pregnancy and risk of autism spectrum disorder in children: A systematic review and meta-analysis of observational studies. Neuroscience and Biobehavioral Reviews. 9 December 2014;49C:82–89. doi:10.1016/j.neubiorev.2014.11.020. PMID 25498856.
- Brown HK, Hussain-Shamsy N, Lunsky Y, Dennis CE, Vigod SN. The Association Between Antenatal Exposure to Selective Serotonin Reuptake Inhibitors and Autism: A Systematic Review and Meta-Analysis.. The Journal of Clinical Psychiatry. January 2017;78(1):e48–e58. doi:10.4088/JCP.15r10194. PMID 28129495.
- Kolevzon A, Gross R, Reichenberg A. Prenatal and perinatal risk factors for autism. Arch Pediatr Adolesc Med. 2007;161(4):326–333. doi:10.1001/archpedi.161.4.326. PMID 17404128.
- Weisskopf MG, Kioumourtzoglou MA, Roberts AL. Air Pollution and Autism Spectrum Disorders: Causal or Confounded?. Current Environmental Health Reports. December 2015;2(4):430–439. doi:10.1007/s40572-015-0073-9. PMID 26399256.
- Flores-Pajot MC, Ofner M, Do MT, Lavigne E, Villeneuve PJ. Childhood autism spectrum disorders and exposure to nitrogen dioxide, and particulate matter air pollution: A review and meta-analysis. Environmental Research. 25 August 2016. doi:10.1016/j.envres.2016.07.030. PMID 27609410.
- Rutter M. Incidence of autism spectrum disorders: changes over time and their meaning. Acta Paediatr. 2005;94(1):2–15. doi:10.1111/j.1651-2227.2005.tb01779.x. PMID 15858952.
- Schultz RT. Developmental deficits in social perception in autism: the role of the amygdala and fusiform face area. Int J Dev Neurosci. 2005;23(2–3):125–41. doi:10.1016/j.ijdevneu.2004.12.012. PMID 15749240.
- Ashwood P, Van de Water J. Is autism an autoimmune disease? Autoimmun Rev. 2004;3(7–8):557–562. doi:10.1016/j.autrev.2004.07.036. PMID 15546805.
- Ashwood P, Wills S, Van de Water J. The immune response in autism: a new frontier for autism research. J Leukoc Biol. 2006 [archived 2006-10-05; Retrieved 2008-02-27];80(1):1–15. doi:10.1189/jlb.1205707. PMID 16698940.
- Stigler KA, Sweeten TL, Posey DJ, McDougle CJ. Autism and immune factors: a comprehensive review. Res Autism Spectr Disord. 2009;3(4):840–860. doi:10.1016/j.rasd.2009.01.007.
- Wills S, Cabanlit M, Bennett J, Ashwood P, Amaral D, Van de Water J. Autoantibodies in autism spectrum disorders (ASD). Ann N Y Acad Sci. 2007;1107:79–91. doi:10.1196/annals.1381.009. PMID 17804535.
- Schmitz C, Rezaie P. The neuropathology of autism: where do we stand? Neuropathol Appl Neurobiol. 2008;34(1):4–11. doi:10.1111/j.1365-2990.2007.00872.x. PMID 17971078.
- Wu S. Family history of autoimmune diseases is associated with an increased risk of autism in children: A systematic review and meta-analysis.. Neuroscience and Biobehavioral Reviews. 15 May 2015;55:322–332. doi:10.1016/j.neubiorev.2015.05.004. PMID 25981892.
- Fox E, Amaral D, Van de Water J. Maternal and fetal antibrain antibodies in development and disease. Dev Neurobiol. 2012;72(10):1327–1334. doi:10.1002/dneu.22052. PMID 22911883.
- Israelyan N, Margolis KG (2018). "Serotonin as a link between the gut-brain-microbiome axis in autism spectrum disorders". Pharmacol Res (Review). 132: 1–6. doi:10.1016/j.phrs.2018.03.020. PMC 6368356. PMID 29614380.
- Wasilewska J, Klukowski M (2015). "Gastrointestinal symptoms and autism spectrum disorder: links and risks - a possible new overlap syndrome". Pediatric Health Med Ther (Review). 6: 153–166. doi:10.2147/PHMT.S85717. PMC 5683266. PMID 29388597.
- Rao M, Gershon MD (September 2016). "The bowel and beyond: the enteric nervous system in neurological disorders". Nat Rev Gastroenterol Hepatol (Review). 13 (9): 517–28. doi:10.1038/nrgastro.2016.107. PMC 5005185. PMID 27435372.
- Azhari A, Azizan F, Esposito G (2018). "A systematic review of gut-immune-brain mechanisms in Autism Spectrum Disorder". Dev Psychobiol (Systematic Review). 61 (5): 752–771. doi:10.1002/dev.21803. PMID 30523646.
- Johnson TW. Dietary considerations in autism: identifying a reasonable approach. Top Clin Nutr. 2006;21(3):212–225. doi:10.1097/00008486-200607000-00008.
- Krishnaswami S, McPheeters ML, Veenstra-Vanderweele J. A systematic review of secretin for children with autism spectrum disorders. Pediatrics. 2011;127(5):e1322–1325. doi:10.1542/peds.2011-0428. PMID 21464196.
- Panksepp J. A neurochemical theory of autism. Trends in Neurosciences. 1979;2:174–177. doi:10.1016/0166-2236(79)90071-7.
- Millward C, Ferriter M, Calver S, Connell-Jones G (2008). "Gluten- and casein-free diets for autistic spectrum disorder". Cochrane Database Syst Rev (Review) (2): CD003498. doi:10.1002/14651858.CD003498.pub3. PMC 4164915. PMID 18425890.
- Shattock P, Whiteley P (2002). "Biochemical aspects in autism spectrum disorders: updating the opioid-excess theory and presenting new opportunities for biomedical intervention". Expert Opin Ther Targets (Review). 6 (2): 175–83. doi:10.1517/14728222.6.2.175. PMID 12223079.
- Christison GW, Ivany K. Elimination diets in autism spectrum disorders: any wheat amidst the chaff?. J Dev Behav Pediatr. 2006;27(2 Suppl 2):S162–171. doi:10.1097/00004703-200604002-00015. PMID 16685183.
- Buie T (2013). "The relationship of autism and gluten". Clin Ther (Review). 35 (5): 578–83. doi:10.1016/j.clinthera.2013.04.011. PMID 23688532.
At this time, the studies attempting to treat symptoms of autism with diet have not been sufficient to support the general institution of a gluten-free or other diet for all children with autism.
- Marí-Bauset S, Zazpe I, Mari-Sanchis A, Llopis-González A, Morales-Suárez-Varela M (December 2014). "Evidence of the gluten-free and casein-free diet in autism spectrum disorders: a systematic review". J Child Neurol. 29 (12): 1718–27. doi:10.1177/0883073814531330. hdl:10171/37087. PMID 24789114.
- Millward C, Ferriter M, Calver S, Connell-Jones G (April 2008). Ferriter M (ed.). "Gluten- and casein-free diets for autistic spectrum disorder". The Cochrane Database of Systematic Reviews (2): CD003498. doi:10.1002/14651858.CD003498.pub3. PMC 4164915. PMID 18425890.
- Volta U, Caio G, De Giorgio R, Henriksen C, Skodje G, Lundin KE (June 2015). "Non-celiac gluten sensitivity: a work-in-progress entity in the spectrum of wheat-related disorders". Best Pract Res Clin Gastroenterol. 29 (3): 477–91. doi:10.1016/j.bpg.2015.04.006. PMID 26060112.
autism spectrum disorders (ASD) have been hypothesized to be associated with NCGS [47,48]. Notably, a gluten- and casein-free diet might have a positive effect in improving hyperactivity and mental confusion in some patients with ASD. This very exciting association between NCGS and ASD deserves further study before conclusions can be firmly drawn
- San Mauro I, Garicano E, Collado L, Ciudad MJ (December 2014). "¿Es el gluten el gran agente etiopatogenico de enfermedad en el siglo XXI?" [Is gluten the great etiopathogenic agent of disease in the XXI century?]. Nutr Hosp (in Spanish). 30 (6): 1203–10. doi:10.3305/nh.2014.30.6.7866. PMID 25433099.
- Kočovská E, Fernell E, Billstedt E, Minnis H, Gillberg C. Vitamin D and autism: clinical review. Res Dev Disabil. 2012;33(5):1541–1550. doi:10.1016/j.ridd.2012.02.015. PMID 22522213.
- Zafeiriou DI, Ververi A, Vargiami E. Childhood autism and associated comorbidities. Brain Dev. 2007;29(5):257–272. doi:10.1016/j.braindev.2006.09.003. PMID 17084999.
- Mehler MF, Purpura DP. Autism, fever, epigenetics and the locus coeruleus. Brain Res Rev. 2009;59(2):388–392. doi:10.1016/j.brainresrev.2008.11.001. PMID 19059284.
- Austin D. An epidemiological analysis of the 'autism as mercury poisoning' hypothesis. Int J Risk Saf Med. 2008;20(3):135–142. doi:10.3233/JRS-2008-0436.
- Nelson KB, Bauman ML. Thimerosal and autism?. Pediatrics. 2003;111(3):674–679. doi:10.1542/peds.111.3.674. PMID 12612255.
- Davidson PW, Myers GJ, Weiss B. Mercury exposure and child development outcomes. Pediatrics. 2004;113(4 Suppl):1023–1029. doi:10.1542/peds.113.4.S1.1023. PMID 15060195.
- Ng DK, Chan CH, Soo MT, Lee RS. Low-level chronic mercury exposure in children and adolescents: meta-analysis. Pediatr Int. 2007;49(1):80–87. doi:10.1111/j.1442-200X.2007.02303.x. PMID 17250511.
- Ng F, Berk M, Dean O, Bush AI. Oxidative stress in psychiatric disorders: evidence base and therapeutic implications. Int J Neuropsychopharmacol. 2008;11(6):851–876. doi:10.1017/S1461145707008401. PMID 18205981.
- Kern JK, Jones AM. Evidence of toxicity, oxidative stress, and neuronal insult in autism. J Toxicol Environ Health B Crit Rev. 2006;9(6):485–499. doi:10.1080/10937400600882079. PMID 17090484.
- Ghanizadeh A, Akhondzadeh S, Hormozi M, Makarem A, Abotorabi-Zarchi M, Firoozabadi A. Glutathione-related factors and oxidative stress in autism, a review. Curr. Med. Chem.. 2012;19(23):4000–4005. doi:10.2174/092986712802002572. PMID 22708999.
- Villagonzalo KA, Dodd S, Dean O, Gray K, Tonge B, Berk M. Oxidative pathways as a drug target for the treatment of autism. Expert Opin. Ther. Targets. 2010;14(12):1301–1310. doi:10.1517/14728222.2010.528394. PMID 20954799.
- Hacking I. The Social Construction of What? Harvard University Press; 1999. ISBN 0-674-00412-4. p. 114–123.
- Nadesan MH. Constructing Autism: Unravelling the 'Truth' and Understanding the Social. Routledge; 2005. ISBN 0-415-32181-6. The dialectics of autism: theorizing autism, performing autism, remediating autism, and resisting autism. p. 179–213.
- Bettelheim B. The Empty Fortress: Infantile Autism and the Birth of the Self. Free Press; 1967. ISBN 0-02-903140-0.
- Kanner L. Autistic disturbances of affective contact. Nerv Child. 1943;2:217–250. Reprinted in Acta Paedopsychiatr. 1968;35(4):100–136. PMID 4880460.
- Kanner L. Problems of nosology and psychodynamics in early childhood autism. Am J Orthopsychiatry. 1949;19(3):416–426. doi:10.1111/j.1939-0025.1949.tb05441.x. PMID 18146742.
- Gardner M. The brutality of Dr. Bettelheim. Skeptical Inquirer. 2000;24(6):12–14.
- Fombonne E, Zakarian R, Bennett A, Meng L, McLean-Heywood D. Pervasive developmental disorders in Montreal, Quebec, Canada: prevalence and links with immunizations. Pediatrics. 2006;118(1):e139–150. doi:10.1542/peds.2005-2993. PMID 16818529.

- Gross L. A broken trust: lessons from the vaccine–autism wars. PLoS Biology. 2009;7(5):e1000114. doi:10.1371/journal.pbio.1000114. PMID 19478850.

- Taylor LE, Swerdfeger AL, Eslick GD. Vaccines are not associated with autism: an evidence-based meta-analysis of case-control and cohort studies. Vaccine. 2014;32(29):3623–3629. doi:10.1016/j.vaccine.2014.04.085. PMID 24814559.

- Hilton S, Petticrew M, Hunt K. 'Combined vaccines are like a sudden onslaught to the body's immune system': parental concerns about vaccine 'overload' and 'immune-vulnerability'. Vaccine. 2006;24(20):4321–4327. doi:10.1016/j.vaccine.2006.03.003. PMID 16581162.

- Gerber JS, Offit PA. Vaccines and autism: a tale of shifting hypotheses. Clinical Infectious Diseases. 2009;48(4):456–461. doi:10.1086/596476. PMID 19128068.

- Paul R. Parents ask: am I risking autism if I vaccinate my children?. Journal of Autism and Developmental Disorders. 2009;39(6):962–963. doi:10.1007/s10803-009-0739-y. PMID 19363650.

- Foster, Craig A.; Ortiz, Sarenna M. (2017). "Vaccines, Autism, and the Promotion of Irrelevant Research: A Science-Pseudoscience Analysis". Skeptical Inquirer. 41 (3): 44–48. Archived from the original on 2018-10-06. Retrieved 6 October 2018.
- Retraction – Ileal-lymphoid-nodular hyperplasia, non-specific colitis, and pervasive developmental disorder in children. Lancet. 2010-02-06;375(9713):445. doi:10.1016/S0140-6736(10)60175-4. PMID 20137807.
- Wakefield A, Murch S, Anthony A et al.. Ileal-lymphoid-nodular hyperplasia, non-specific colitis, and pervasive developmental disorder in children. Lancet. 1998;351(9103):637–641. doi:10.1016/S0140-6736(97)11096-0. PMID 9500320. (Retracted, see doi:10.1016/S0140-6736(10)60175-7)
- Murch SH, Anthony A, Casson DH et al. Retraction of an interpretation. Lancet. 2004;363(9411):750. doi:10.1016/S0140-6736(04)15715-2. PMID 15016483.
- Deer B. The MMR-autism crisis – our story so far; 2008-11-02 [Retrieved 2008-12-06].
- Centers for Disease Control and Prevention. Measles, mumps, and rubella (MMR) vaccine; 2008-12-23 [Retrieved 2009-02-14].
- Institute of Medicine, National Academy of Sciences. Immunization safety review: vaccines and autism; 2004 [archived 2007-06-23; Retrieved 2007-06-13].
- National Health Service. MMR the facts [archived 2007-06-15; Retrieved 2007-06-13].
- Godlee F, Smith J, Marcovitch H. Wakefield's article linking MMR vaccine and autism was fraudulent. BMJ. 2011;342:c7452. doi:10.1136/bmj.c7452. PMID 21209060.
- Deer B. How the case against the MMR vaccine was fixed. BMJ. 2011;342:c5347. doi:10.1136/bmj.c5347. PMID 21209059.
- Study linking vaccine to autism was fraud. 2011-01-05 [archived 2011-01-07; Retrieved 2011-01-06]. Associated Press. NPR.
- Retracted autism study an 'elaborate fraud,' British journal finds. (Atlanta) 2011-01-06 [Retrieved 2011-01-06].
- "Vaccines, blood and biologics: thimerosal in vaccines". US Food and Drug Administration. 2012. Retrieved October 24, 2013.
- Eaton L. Measles cases in England and Wales rise sharply in 2008. BMJ. 2009;338:b533. doi:10.1136/bmj.b533. PMID 19208716.
- Choi YH, Gay N, Fraser G, Ramsay M. The potential for measles transmission in England. BMC Public Health. 2008;8:338. doi:10.1186/1471-2458-8-338. PMID 18822142.
- https://sciencebasedmedicine.org/mercury-in-vaccines-and-autism-a-failed-hypothesis/
- American Medical Association. AMA Welcomes New IOM Report Rejecting Link Between Vaccines and Autism; 2004-05-18 [Retrieved 2007-07-23].
- American Academy of Pediatrics. What Parents Should Know About Thimerosal; 2004-05-18 [archived 2007-07-08; Retrieved 2007-07-23].
- Kurt TL. ACMT position statement: the Iom report on thimerosal and autism [PDF]. J Med Toxicol. 2006 [archived 2008-02-29; Retrieved 2009-04-12];2(4):170–171. doi:10.1007/BF03161188. PMID 18072140.
- Infectious Diseases and Immunization Committee, Canadian Paediatric Society. Autistic spectrum disorder: No causal relationship with vaccines. Paediatr Child Health. 2007 [archived 2008-12-02; Retrieved 2008-10-17];12(5):393–395. Also published (2007) in Can J Infect Dis Med Microbiol 18 (3): 177–179. PMID 18923720.
- "Thimerosal in vaccines". Center for Biologics Evaluation and Research, U.S. Food and Drug Administration. 2007-09-06. Retrieved 2007-10-01.
- World Health Organization (2006). "Questions and answers about autism spectrum disorders (ASD)". Retrieved 2014-11-02.
- National Advisory Committee on Immunization. Thimerosal: updated statement. An Advisory Committee Statement. Can Commun Dis Rep. 2007;33(ACS-6):1–13. PMID 17663033.
- European Medicines Agency. EMEA Public Statement on Thiomersal in Vaccines for Human Use; 2004-03-24 [archived 2007-06-10; Retrieved 2007-07-22].