Chapter 11: Antimicrobial Susceptibility Testing of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae
Printer friendly version [48 pages]
Each laboratory must decide their own level of susceptibility testing to provide the essential data for public health decision making relevant to that laboratory’s situation. N. meningitidis, H. influenzae, and S. pneumoniae have all been associated with treatment or chemoprophylaxis failures due to strains resistant to or with reduced susceptibility to the antimicrobials used. In addition to monitoring for clinical or chemoprophylactic failures, surveillance of the antibiotic susceptibility patterns in circulating strains of N. meningitidis, H. influenzae, and S. pneumoniae is part of monitoring the emergence and spread of strains with reduced susceptibility to antimicrobials. In order for a laboratory to successfully undertake isolation, identification, and antimicrobial susceptibility testing responsibilities, it must participate in on-going investments in materials, supplies, media, reagents, and quality control, along with periodic training of personnel and quality assessment or proficiency testing. Any deviations from antimicrobial susceptibility testing methods as described in the following pages may invalidate the test results, especially for fastidious organisms such as N. meningitidis, H. influenzae, and S. pneumoniae.
Antimicrobial susceptibility test methods must be performed as described according to internationally recognized clinical guidelines such as those provided by the Clinical and Laboratory Standards Institute (CLSI) (formerly known as National Committee on Clinical Laboratory Standards – NCCLS) (http://www.clsi.org/), which is an international, interdisciplinary, nonprofit, educational organization that develops updated consensus standards and guidelines for the healthcare community on an annual basis. The Comité de l’Antibiogramme de la Société Française de Microbiologie (CA-SFM) (http://www.sfm-microbiologie.org/) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/), whose main objectives are to harmonize breakpoints for antimicrobial agents in Europe, to act as the breakpoint committee for the European Medicines Agency (EMEA) during the registration of new antimicrobial agents, and to also provide internationally recognized clinical guidelines. The overarching goals of these committees are to provide meaningful guidelines for clinical and epidemiological interpretation of results.
There are a variety of methods by which one can determine the antimicrobial susceptibility of a bacterial pathogen, commonly including disk diffusion, agar dilution or broth microdilution, and antimicrobial gradient strip diffusion (14). The disk diffusion method presented in this chapter is a modification of the Kirby-Bauer technique that has been carefully standardized by CLSI and others. If performed precisely according to the following protocol, this method will yield data that can reliably predict the in vivo effectiveness of the drug in question. Although disk diffusion will provide information for most antimicrobial agents regarding interpretation of a strain as susceptible, intermediate, or resistant, it does not provide accurate information about the minimal inhibitory concentration (MIC). In addition, disk diffusion does not produce reliable results with some antibiotic/organism combinations, such as for penicillin G in N. meningitidis and S. pneumoniae. Therefore, this laboratory manual also recommends use of antimicrobial gradient strip diffusion to gather data about the MIC of antimicrobial agents.
Antimicrobial gradient strips are an antimicrobial susceptibility testing method that is as technically simple to perform as disk diffusion and produces semi-quantitative results that are measured in micrograms per milliliter (µg/ml). It is drug-specific, consists of a thin plastic antibiotic gradient strip that is applied to an inoculated agar plate, and is convenient in that it applies the principles of agar diffusion to perform semi-quantitative testing. The continuous concentration gradient of stabilized, dried antibiotic is equivalent to 15 two-fold dilutions by a conventional reference MIC procedure as suggested by CLSI. Antimicrobial gradient test strips have been compared and evaluated beside both the agar and broth dilution susceptibility testing methods recommended by CLSI. Authoritative reports indicate that an ~85-100% correlation exists between the accepted conventional MIC determinations and the MIC determined by the test strip procedure for a variety of organism-drug combinations (2, 3, 9, 10). Some studies have cited gradient test strip MICs as approximately one dilution higher than MICs determined by standard dilution methods.
MIC testing can also be done by dilution; but because agar dilution and broth microdilution are expensive and technically complex, this manual recommends that countries that do not currently do MIC testing by dilution methods should utilize a reference laboratory rather than developing the assay in-country. Alternatively, if resources are available, laboratories may purchase commercially available, frozen MIC panels and follow the manufacturer’s instructions to carry out the MIC test. It is important to note that the accuracy and reproducibility of these tests are dependent on following standard quality control/quality assurance (QC/QA) testing procedures and conditions in laboratories on an on-going basis.
This guide describes the optimal media, inoculum, antimicrobial agents to test, incubation conditions, and interpretation of results for N. meningitidis, H. influenzae, and S. pneumoniae put forth by CLSI, CA-SFM, and EUCAST. In multiple instances, the zone diameter and MIC interpretative standards differ for the same antimicrobial between CLSI, CA-SFM, and EUCAST. These differences arise for many reasons, including: different databases of susceptibility data, differences in interpretation of that data, differences in both antimicrobials and dosages used in different parts of the world, and public health policies. The interpretive standards put forth by all 3 organizations are to be treated as guidelines and may be modified to meet the needs of the region. It is incumbent on the laboratory and public health system to remain alert for clinical treatment failures and trends of decreasing susceptibility to antimicrobials, regardless of which set of interpretive standards are utilized.
Top of Page- Antimicrobial susceptibility program recommendations
Antimicrobial susceptibility testing is a resource-intensive activity requiring a significant amount of labor, well-trained technicians, and quality control processes that must be maintained. Each laboratory considering starting a testing program should perform a cost-benefit analysis to determine the amount of testing that can be done without adversely affecting other laboratory functions. While the optimal testing situation would be to perform susceptibility testing on all incoming isolates, that is unlikely to be practical or economical. Susceptibility testing of a subset of both endemic and epidemic isolates (i.e., every 10th isolate) would provide useful data. During an epidemic caused by a clonal strain, testing every 25th isolate may be sufficient.
These numbers are arbitrary and may have to be revised as the epidemiologic situation changes. If an isolate is found to be resistant to a given antimicrobial, it would be prudent to test more isolates epidemiologically associated with the resistant isolate.
It is imperative that monitoring for clinical and/or chemoprophylaxis failures be performed regardless of the amount of susceptibility testing being performed. A communication network should be set up to allow clinicians to notify public health officials of the potential treatment failure and to ship specimens from suspected treatment or chemoprophylaxis failures to the reference laboratory for susceptibility testing. A mechanism must also exist to allow clinicians and public health officials to receive the susceptibility data in a timely fashion. In addition, a communication network should include links to pharmacies and pharmacists to monitor for changes in prescription practices and antibiotic usage. Changes may reflect treatment and/or chemoprophylaxis failures and may warrant further investigation.
- Quality control for antimicrobial susceptibility testing of N. meningitidis, H. influenzae, and S. pneumoniae
In order to ensure the validity and accuracy of the results obtained by susceptibility testing, it is vital that a quality control (QC) system be in place in the laboratory. The goals of QC are to verify the repeatability and accuracy of the susceptibility test being used, the performance of reagents used in the tests, and the performance of the laboratorians performing the tests and reading the results. Therefore, it is vital to include control organisms with known zone diameters or MIC ranges to the antibiotics being tested. CLSI, CA-SFM, and EUCAST have recommended strains that are to be used as quality controls for antimicrobial susceptibility tests. See Tables 1-5 for strains and limits for both disk diffusion and MIC determination recommended by CLSI, CA-SFM, and EUCAST. A laboratory should choose which QC strain(s) to use based on the antimicrobials to be tested for susceptibility.
If QC testing of antimicrobial tests are performed daily for 20 or 30 days for each strain and antimicrobial agent combination with no more than 1 out of 20 tests outside of control limits (see Tables 1-5), then the tests can be performed once per week. Alternatively, if testing is done less frequently, then QC testing should be performed with every group of tests. They should also be done with each new batch of antimicrobial susceptibility test medium and every time a new lot of disks or gradient strips are used. Note that CLSI QC and breakpoint guidelines can be found in the document: Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement (5).
-
Corrective action for out of range quality control results
Adapted from:
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard—Tenth Edition. CLSI document M2-A10. Wayne, PA: Clinical and Laboratory Standards Institute; 2009, p 27-33.
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard—Tenth Edition. CLSI document M2-A10. Wayne, PA: Clinical and Laboratory Standards Institute; 2009, p 27-33.
QC results periodically will be out of the normal range. If zone diameters or MICs produced by the control strains are out of the expected ranges, the laboratorian should consider the following possible sources of error:
- Antimicrobial susceptibility tests are affected by variations in media, inoculum size or growth phase, incubation time, temperature, and other environmental factors. The medium used may be a source of error if it fails to conform to CLSI, CA-SFM, or EUCAST recommended guidelines. For example, agar containing excessive amounts of thymidine or thymine can reverse the inhibitory effects of sulfonamides and trimethoprim, causing the zones of growth inhibition to be smaller or less distinct. Organisms may appear to be resistant to these drugs when in fact they are not. QC/QA guidelines for preparation of the media must be closely followed.
- If the depth of the agar in the plate is not uniformly 3-4 mm, the rate of diffusion of the antimicrobial agents or the activity of the drugs may be affected.
- If the pH of the test medium is not between 7.2 and 7.4, the rate of diffusion of the antimicrobial agents or the activity of the drugs may be affected. Note: do not attempt to adjust the pH of the Mueller-Hinton agar test medium if it is outside the range.
Table 1. CLSI recommendations for acceptable limits for quality control strains used to monitor accuracy of Kirby-Bauer disk diffusion
Copyrighted material used with permission from the Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, PA, USA 19087, www.clsi.org.
CLSI document M100-S21; 2011, pp 114-117.
Antimicrobial
AgentDisk
ContentE. coli1
ATCC 259222
zone diameter, nearest whole mmH. influenzae3
ATCC 49427
zone diameter, nearest whole mmS. pneumoniae4
ATCC 49619
zone diameter, nearest whole mmAmpicillin 10 µg 16-22 13-21 30-36 Ceftriaxone 30 µg 29-35 31-39 30-35 Ciprofloxacin 5 µg 30-40 34-42 -5 Erythromycin 15 µg - - 25-30 Penicillin 10 units - - 24-30 Rifampicin 5 µg 8-10 22-30 25-30 Trimethoprim-sulfamethoxazole (cotrimoxazole)6 1.25/23.75 µg 23-29 24-32 20-28
Footnotes
1Values are valid for testing this QC strain on Mueller-Hinton agar without blood or other supplements.
2ATCC: American Type Culture Collection, Manassas, Virginia, USA. http://www.atcc.org/.
3Values are valid for testing this QC strain on Haemophilus test medium incubated in 5% CO2 for 16-18 hr at 35°C.
4Values are valid for testing this QC strain on Mueller-Hinton agar supplemented with 5% defibrinated sheep blood incubated in 5% CO2 for 16-18 hr at 35°C.
5- = no data available.
6Trimethoprim-sulfamethoxazole is a combination of two drugs used in treatment (cotrimoxazole). The 1:20 ratio is that at which the greatest synergy in treatment has been demonstrated in serum. The disks are impregnated with 1.25 µg trimethoprim and 23.75 µg sulfamethoxazole to mimic the 1:20 ratio.
Table 2. CLSI recommendations for acceptable limits for QC strains used to monitor accuracy of minimal inhibitory concentrations (MIC)
Copyrighted material used with permission from the Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, PA, USA 19087, www.clsi.org.
CLSI document M100-S21; 2011, pp 122-125.
Antimicrobial
AgentE. coli1
ATCC 25922
(µg/ml)H. influenzae2
ATCC 49427
(µg/ml)S. pneumoniae3
ATCC 49619
(µg/ml)Ampicillin 2-8 2-8 0.06-0.25 Ceftriaxone 0.03-0.12 0.06-0.25 0.03-0.12 Ciprofloxacin 0.004-0.0154 0.004-0.03 -5 Erythromycin - - 0.03-0.12 Penicillin - - 0.25-1 Rifampicin 4-16 0.25-1 0.015-0.06 Tetracycline 0.5-2 4-32 0.06-0.5 Trimethoprim-sulfamethoxazole (cotrimoxazole)6 ≤0.5/9.5 0.03/0.59-0.25/4.75 0.12/2.4-1/19
Footnotes
1Values are valid for testing this QC strain on cation-adjusted Mueller-Hinton broth (CAMHB) without blood or other supplements.
2Values are valid for testing this QC strain on Haemophilus test medium broth incubated in ambient air for 20-24 hr at 35°C.
3Values are valid for testing this QC strain on CAMHB with lysed horse blood (2.5-5.0% v/v) incubated in 5% CO2 or ambient air for 20-24 hr at 35°C. Note that for S. pneumoniae that is not ATCC 49619, the plates should be incubated in ambient air.
4QC limits are the same for ciprofloxacin if E. coli ATCC 25922 is tested in CAMHB with lysed horse blood (2.5-5.0% v/v).
5- = no data available.
6Trimethoprim-sulfamethoxazole is a combination of two drugs used in treatment (cotrimoxazole). The greatest synergy in serum has been found when the two drugs are at a 1:20 ratio, thus the breakpoints are given at a 1/20 ratio of trimethoprim/sulfamethoxazole; i.e., ≤0.5 µg/ml trimethoprim/9.5 µg/ml sulfamethoxazole.
Table 3. CA-SFM recommendations for acceptable limits for QC strains used to monitor accuracy of Kirby-Bauer disk diffusion and to monitor accuracy of MIC
Kirby-Bauer disk diffusion QC limits are given for E. coli CIP 7624 and S. aureus CIP 7625 and MIC QC limits are given for S. pneumoniae CIP 107808. Many antibiotics listed in this chapter do not have QC limits set by CA-SFM.
Comite de L’Antibiogramme de la Société Française de Microbiologie, Société Française de Microbiologie, Recommendations 2010, p8.
Antimicrobial
AgentDisk
ContentE. coli1
CIP 76242
zone diameter, nearest whole mmStaphylococcus aureus1
CIP 76253
zone diameter, nearest whole mmS. pneumoniae4
CIP 104485
(µg/ml)Cefotaxime 30 µg 32.5-37.5 - 5 0.03-0.25 Ciprofloxacin 5 µg 31-38 - - Erythromycin 15 units - 26.5-31.5 - Penicillin G 6 µg
(10 units)- 31-38.5 0.125-0.5 Rifampicin 30 µg - 34.0-39.0 - Trimethoprim-sulfamethoxazole (cotrimoxazole)6 1.25/23.75 µg 25.5-30.5 28.0-32.5 -
Footnotes
1Values are valid for testing this QC strain on Mueller-Hinton agar, McFarland 0.5, incubated in ambient air for 20-24 h at 35-37°C.
2CIP: Collection de Bactéries de l´ Institut Pasteur, Paris, France http://www.crbip.pasteur.fr/. This strain is equivalent to E. coli ATCC 25922.
3CIP: This strain is equivalent to S. aureus ATCC 25923.
4Values are valid for testing this QC strain on Mueller-Hinton agar supplemented with 5% sheep blood, McFarland 0.5, incubated in ambient air for 18-24 h at 35-37°C. For cotrimoxazole, use Mueller-Hinton agar supplement with 5% haemolysed horse blood.
5- = no data available.
6Trimethoprim-sulfamethoxazole is a combination of two drugs used in treatment (cotrimoxazole). The 1:20 ratio is that at which the greatest synergy in treatment has been demonstrated in serum. The disks are impregnated with 1.25 µg trimethoprim and 23.75 µg sulfamethoxazole to mimic the 1:20 ratio.
Table 4. EUCAST recommendations for acceptable limits for QC strains used to monitor accuracy of Kirby-Bauer disk diffusion
Data is from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) website, http://www.eucast.org, version 1.3, December 2010. More information can be found at this website.
Antimicrobial
AgentDisk
ContentE. coli1
ATCC 259222
zone diameter, nearest whole mmH. influenzae3
ATCC 93344
zone diameter, nearest whole mmS. pneumoniae5
ATCC 496196
zone diameter, nearest whole mmAmpicillin 2 µg - 7 19-25 25-31 Ampicillin 10 µg 16-22 - - Azithromycin 15 µg - - - Benzylpenicillin 1 unit - - 15-21 Cefotaxime 5 µg 25-31 29-35 28-34 Ceftriaxone 30 µg 29-35 33-41 32-38 Chloramphenicol 30 µg 21-27 30-38 24-30 Ciprofloxacin 5 µg 30-40 31-39 22-28 Clindamycin 2 µg - - 22-28 Erythromycin 15 µg - 12-18 26-32 Levofloxacin 5 µg 29-37 32-38 21-27 Meropenem 10 µg 28-34 28-34 30-38 Nalidixic acid 30 µg 22-28 27-33 - Rifampicin 5 µg - 20-26 26-32 Telithromycin 15 µg - 15-21 27-33 Tetracycline 30 µg 18-25 28-34 28-34 Trimethoprim-sulfamethoxazole (cotrimoxazole)8 1.25/23.75 µg 23-29 26-34 20-26
Footnotes
1Values are valid for testing this QC strain on Mueller-Hinton agar, McFarland 0.5, air, 35±1ºC, 18±2 hours.
2Equivalent E. coli QC strains are: NCTC 12241, CIP 7624, DSM 1103, and CCUG 17620 NCTC: National Collection of Type Cultures, PHLS Central Public Health Laboratory, London, U.K. http://www.hpacultures.org.uk/collections/nctc.jsp CIP : Collection de Bactéries de l´ Institut Pasteur, Paris, France http://www.crbip.pasteur.fr/ DSM: DSMZ Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany http://www.dsmz.de/ CCUG: Culture Collection, University of Göteborg, Department of Clinical Bacteriology, Göteborg, Sweden http://www.ccug.se/
3Values are valid for testing this quality control strain on Mueller-Hinton agar with 5% horse blood and 20 µg/ml NAD, McFarland 0.5, 5% CO2, 35±1ºC, 18±2 hours.
4Equivalent H. influenzae QC strains are: NCTC 8468, CIP 54.94, and CCUG 23946.
5Values are valid for testing this QC strain on Mueller-Hinton agar with 5% horse blood and 20 µg/ml b-NAD, McFarland 0.5, 5% CO2, 35±1°C, 18±2 hours.
6Equivalent S. pneumoniae QC strains are: NCTC 12977, CIP 104340, DSM 11967, and CCUG 33638.
7- = no data available.
8Trimethoprim-sulfamethoxazole is a combination of two drugs used in treatment (cotrimoxazole). The 1:20 ratio is that at which the greatest synergy in treatment has been demonstrated in serum. The disks are impregnated with 1.25 µg trimethoprim and 23.75 µg sulfamethoxazole to mimic the 1:20 ratio.
Table 5. EUCAST recommendations for acceptable limits for QC strains used to monitor accuracy of MIC*
Data is from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) website, http://www.eucast.org, version 1.3, December 2010. More information can be found at this website.
*EUCAST does not yet have established MIC QC limits for H. influenzae ATCC 9334
Antimicrobial
AgentE. coli1
ATCC 259222
(µg/ml)S. pneumoniae3
ATCC 496194
(µg/ml)Ampicillin 2-8 0.064-0.25 Azithromycin -5 0.064-0.25 Benzylpenicillin - -0.25-1 Cefotaxime 0.032-0.125 0.032-0.125 Ceftriaxone 0.032-0.125 0.032-0.125 Chloramphenicol 2-8 2-8 Ciprofloxacin 0.004-0.016 - Clindamycin - 0.032-0.125 Erythromycin - 0.032-0.125 Levofloxacin 0.008-0.064 0.5-2 Meropenem 0.008-0.064 0.064-0.25 Nalidixic acid 1-4 - Rifampicin - 0.016-0.064 Telithromycin 0.004-0.032 Tetracycline - 0.125-0.5 Trimethoprim-sulfamethoxazole (cotrimoxazole)6 ≤0.5/9.57 0.125/2.4-1/19
Footnotes
1Values are valid for testing this QC strain on Mueller-Hinton agar, McFarland 0.5, air, 35±1°C, 18±2 hours.
2Equivalent E. coli QC strains are: NCTC 12241, CIP 76.24, DSM 1103, and CCUG 17620.
3Values are valid for testing this QC strain on Mueller-Hinton agar with 5% horse blood and 20 µg/ml b-NAD, McFarland 0.5, 5% CO2, 35±1°C, 18±2 hours.
4Equivalent S. pneumoniae QC strains are: NCTC 12977, CIP 104340, DSM 11967, and CCUG 33638.
5- = no data available.
6Trimethoprim-sulfamethoxazole is a combination of two drugs used in treatment (cotrimoxazole). The greatest synergy in serum has been found when the two drugs are at a 1:20 ratio, thus the breakpoints are given at a 1/20 ratio of trimethoprim/sulfamethoxazole; i.e., ≤0.12 µg/ml trimethoprim/2.4 µg/ml sulfamethoxazole.
7Target set by International Standard ISO 20776-1: 2006. No range available from EUCAST.
- If the inoculum is not a pure culture or does not contain a concentration of bacteria that approximates the 0.5 McFarland turbidity standard, the antimicrobial susceptibility test results will be affected. For instance, a resistant organism could appear to be susceptible if too few bacteria are used in the inoculum. Also, even if the isolates are susceptible, when colonies from blood agar medium are used to prepare a suspension by the direct inoculum method, trimethoprim or sulfonamide antagonists may be carried over and produce a haze of growth inside the zones of inhibition surrounding trimethoprim-sulfamethoxazole (cotrimoxazole) disks. In addition, only cultures in growth phase, i.e., cultures grown within 20-24 hours, should be used.
If the out of range result is due to an obvious reason such as use of the wrong disk or gradient strip, use of the wrong control strain, obvious contamination of the strain, or use of the wrong incubation temperatures or conditions, then document the reason and retest the strain on the day the error is observed. If the repeated result is within range, no further corrective action is required.
If no obvious reason for the out of range result is apparent, immediate corrective action is required. Test the out of range antimicrobial agent/organism combination on the day the error is observed and monitor for a total of five consecutive test days and document all results. If the results of the tests from all five days are within acceptable range, no additional corrective action is necessary. However, if any of the five results remain out of range, then additional corrective action is required. In the interim, daily control tests must be performed until the problem is solved.
If immediate corrective action does not solve the problem, then other common sources of error need to be investigated to verify that:
- Results were measured and transcribed correctly.
- The turbidity standard had not expired, was stored properly, met performance requirements, and was adequately mixed prior to use.
- All materials, including disks and gradient test strips, used were within their expiration dates and were stored and used at the proper temperature.
- The incubator was at the proper temperature and atmosphere.
- Other equipment used such as pipettors were functioning properly and measuring accurately.
- The control strain had not changed and was not contaminated.
- Inoculum suspensions were prepared and adjusted correctly.
- The inoculum was used within 15 minutes of preparation.
- Inoculum for the test was prepared from a plate incubated for the correct length of time and was not more than 24 hours old.
If necessary, obtain a new QC strain from either freezer storage or from a reliable source. New lots of materials may be necessary, as well. It may be helpful to exchange quality control strains and materials with another laboratory using the same method to verify results. Until the problem is resolved, it may be necessary to use an alternative test method, if one is available.
-
- Antimicrobial susceptibility testing of N. meningitidis
N. meningitidis does not commonly show widespread resistance to many antimicrobial agents, though resistance to sulfonamides has become quite common (8). N. meningitidis with reduced susceptibility to penicillin is common in many areas of the world, though the clinical significance of this resistance has not yet been established (10, 13, 22). In addition, resistance to rifampicin has been reported and such strains have resulted in prophylaxis and treatment failures (16, 19, 21). Recently, sporadic resistance to ciprofloxacin, an antibiotic commonly used for chemoprophylaxis of non-pregnant adults in many countries, has been reported throughout the world, including Europe, South America, Australia, Asia, and North America (1, 4, 6, 18, 20, 23). Third generation cephalosporins such as ceftriaxone tend to be the empiric drugs of choice for primary treatment in most areas of the world but ceftriaxone non-susceptible isolates have been reported in India (12).
Susceptibility testing of N. meningitidis is difficult as a consensus has not been reached regarding the best techniques for N. meningitidis susceptibility testing or their standardization. The guidelines presented here must be adapted to each laboratory’s needs, capabilities, and capacities as well as to the overarching public health requirements. This manual contains suggested antibiotics to screen for reduced susceptibility and guidelines on how to both perform and interpret susceptibility tests, but they should be modified and interpreted to meet the needs of each country, region, and laboratory. CLSI, CA-SFM, EUCAST, and other international organizations are working together to attempt to reach consensus recommendations.
-
Selection of antibiotics to screen for susceptibility
Selecting the antibiotics to screen for susceptibility is largely driven by the geographical regions in which the isolates originate and the capacity of the laboratory. It is recommended to routinely determine the susceptibility of circulating strains of N. meningitidis against the antimicrobials used for primary treatment and chemoprophylaxis in the geographical region from which the specimens originate. It may not be necessary for the laboratory to perform susceptibility testing on all routine surveillance isolates; a random representative subset can be tested. Furthermore, during an outbreak it is not necessary to perform susceptibility testing on all isolates. Laboratories could also consider periodic, non-routine surveillance for characteristics such as β-lactamase production in penicillin or cephem class resistant strains, and ceftriaxone and chloramphenicol resistance (if not already routinely tested), for example. These data would help provide information to public health agencies and international reference laboratories regarding the emergence of new N. meningitidis strains with reduced susceptibility to antimicrobials of clinical and public health concern.
-
Biosafety precautions
Several fatal laboratory-acquired meningococcal disease cases have been reported, thus biosafety should be a top priority (17). Biosafety Level 2 (BSL2) practices are recommended for N. meningitidis in most countries. Whenever possible, procedures likely to generate aerosols should be performed in a biological safety cabinet. Consider vaccination of laboratorians that work with invasive meningococcal isolates, if possible, although current vaccines are not protective against all serogroups.
-
Antimicrobial susceptibility testing of N. meningitidis by Kirby-Bauer disk diffusion
Kirby-Bauer disk diffusion is the least expensive screen for antimicrobial susceptibility testing, but results can be difficult to interpret. These tests can be useful for screening isolates to categorize them as susceptible, intermediate, resistant, or non-susceptible for several antimicrobials. However, since this test does not determine MICs, it is not useful for detecting subtle trends of decreasing susceptibility. Kirby-Bauer disk diffusion tests do not produce reliable results for ampicillin and penicillin and false intermediate, resistant, or non-susceptible results are seen with N. meningitidis. False intermediate or resistant results are not unusual when testing the susceptibility of an isolate of N. meningitidis to ciprofloxacin with a 5 µg ciprofloxacin disk. Thus, results demonstrating an isolate with reduced susceptibility should be verified using a MIC test.
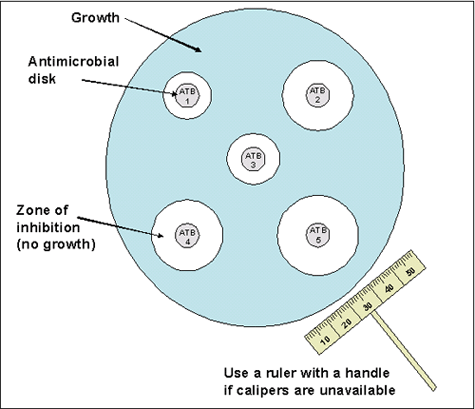
Either 150-mm or 100-mm plates can be used for Kirby-Bauer disk diffusion depending on the number of antimicrobial agents to be tested per isolate. CLSI guidelines state that no more than two disks can be used on a 100-mm plate and up to five disks can be used on a 150-mm plate (Figure 1). Mueller-Hinton with 5% sheep blood agar with a depth of 4 mm is used when testing N. meningitidis isolates using this method.
Isolates to be tested should be subcultured onto a chocolate agar plate and incubated in a CO2-enhanced atmosphere (5% CO2 in a CO2-incubator or candle-extinction jar) at 35±2°C for 20-24 hours prior to testing. If the organism has been frozen, it should be subcultured twice when it is removed from the freezer before proceeding with susceptibility testing.
Remove agar plates from the refrigerator and allow them to come to room temperature (25°C) before inoculating. Warm the cation-adjusted Mueller-Hinton broth (CAMHB) to 35°C before using. Allow the antibiotic disks that will be used in the batch of testing to warm to room temperature (25°C).
-
Using a sterile cotton-tip applicator, touch the surface of one to four morphologically identical, isolated colonies. Immerse the applicator into a tube containing sterile CAMHB. Rub the applicator against the wall of the tube slightly to release a small amount of growth into the liquid. Cap the tube and mix the cells using a vortex to form a suspension, being careful not to form froth or bubbles in the suspension when mixing the cells. Adjust the turbidity of the inoculum to that of a 0.5 McFarland turbidity standard (approximately 1 to 4 x 108 CFU/ml). Preparation of aMcFarland turbidity standard is described inthe Annex. If the turbidity of the inoculum is greater than the standard, dilute it with CAMHB to equal the turbidity of the standard. This suspension must be used within 15 minutes.
- Perform regular colony counts to verify that the density of the inoculum suspension is correct. For example, dilute the suspension 1:100 and subculture 10 µl onto the recommended media. An acceptable inoculum should give approximately 100-500 colonies. It is not necessary to perform colony counts on every isolate tested.
- Immerse a sterile cotton-tipped swab into the adjusted inoculum. Remove excess liquid by pressing the swab tip against the inside of the tube. Inoculate the entire surface of a blood agar plate three times with the same swab of inoculum, rotating the plate 60 degrees after each inoculation to ensure even distribution of the inoculum and confluent growth of the bacteria. Use a single swab of inoculum and do not return the swab to the broth after each rotation.
- Allow the inoculum to dry on the surface of the plate (which should take approximately 5-10 minutes). Be sure the plate is entirely dry before proceeding, but do not exceed 15 minutes.
- When the surface of the inoculated plate is dry and the disks are at room temperature, place the disks onto the agar with an applicator or sterile forceps. Make sure that the disks are spaced enough distance apart on the agar so the zones of inhibition do not overlap (Figure 1). Press down on the disks to ensure complete contact with the agar surface. Alternatively, a mechanical disk dispenser can be used. Once applied, it is important to not move the antibiotic disks as the antibiotic will begin to diffuse immediately upon contact with the plate.
- Incubate the plates in an inverted position in a 5% CO2 atmosphere or candle jar for 20-24 hours at 35±2°C.
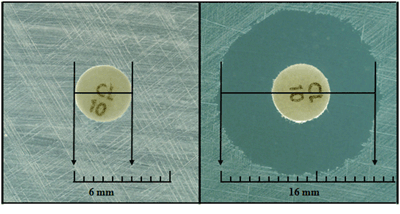
- After overnight incubation, measure the diameter of each zone of inhibition with a ruler or calipers (Figure 2). Measurements should be performed in a biosafety cabinet, if possible. The zones of inhibition on the media containing blood are measured from the top surface of the plate with the top removed. Use either calipers or a ruler with a handle attached for these measurements, holding the ruler over the center of the surface of the disk when measuring the inhibition zone (Figures 1 and 2).
- Care should be taken not to touch the disk or surface of the agar. Decontaminate the ruler occasionally to prevent transmission of the bacteria. In all measurements, the zones of inhibition are measured as the diameter from the edges of the last visible colony of the unaided eye. Record the results to the nearest millimeter (mm).
- Interpret the antimicrobial susceptibility of the strain being tested (and check that results for the QC strains are within the acceptable control range) by comparing the results to the CLSI (Table 6) or CA-SFM (Table 7) standard zone sizes. See Figure 3 for a sample worksheet for recording antimicrobial susceptibility test results for N. meningitidis.
Date of Testing:
Test performed by:
Interpretation of susceptibility: S= susceptible I= intermediate R= resistantSpecimen
numberMeningitis
Isolate?Organism Antibiotic #1 Antibiotic #2 Antibiotic #3 Antibiotic #4 mm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I Rmm
µg/ml
S I RQC strain N/A QC strain mm
µg/ml
Yes Nomm
µg/ml
Yes Nomm
µg/ml
Yes Nomm
µg/ml
Yes NoFigure 3. Sample sheet for recording data and quality control information. Note: After 20-24 hours of incubation, check the results for the QC strain(s) against the standard acceptable ranges; if they are within control limits, continue reading results for the test isolate. Record disk diffusion results in mm and MIC results in µg/ml. Inhibition zone ranges and breakpoints for interpretation of results may be found in Tables 1-5, depending on the QC strains used and the guidelines being followed.
Table 6. Kirby-Bauer disk diffusion zone diameter interpretive standards and MIC interpretive standards for N. meningitidis as recommended by CLSI
Copyrighted material used with permission from the Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, PA, USA 19087, http://www.clsi.org.
CLSI document M100-S21; 2011, pp 108-110.
Antimicrobial
Agent/ClassDisk
ContentZone Diameter,
nearest whole mmMIC Interpretive
Standard (µg/ml)S I R S I R PENICILLINS Penicillin1 - 2 - - ≤ 0.06 0.12-0.25 ≥ 0.5 CEPHEMS Ceftriaxone3 30 µg ≥ 34 - - ≤ 0.12 - - MACROLIDES Azithromycin3,4,5 15 µg ≥ 20 - - ≤ 2 - - FLUOROQUINOLONES Ciprofloxacin4 5 µg ≥ 35 33-34 ≤ 32 ≤ 0.03 0.06 ≥ 0.12 PHENICOLS Chloramphenicol6 30 µg ≥ 26 20-25 ≤ 19 ≤ 2 4 ≥ 8 ANSAMYCINS Rifampicin3 5 µg ≥ 25 20-24 ≤ 19 ≤ 0.5 1 ≥ 2
Footnotes
1Disk diffusion tests with penicillin for N. meningitidis are unreliable. MIC tests should be used for this organism.
2- = no data available.
3For some antimicrobial agents, the absence or rare occurrence of resistant strains precludes defining any results categories other than “susceptible”.
4May be appropriate only for prophylaxis of meningococcal case contacts. These breakpoints do not apply to therapy of patients with invasive meningococcal disease. Recently, it has been suggested that screening for reduced susceptibility to ciprofloxacin using a 30 ug nalidixic acid disk is useful for detecting meningococcal quinolone-resistant strains associated with gyrA mutations (7).
5Interpretive criteria were developed initially using MICs determined by incubation in ambient air for the pharmacodynamic calculations.
6Not routinely reported on isolates from the urinary tract.
Table 7. Kirby-Bauer disk diffusion zone diameter interpretive standards and MIC interpretive standards for N. meningitidis as recommended by CA-SFM
Comite de L’Antibiogramme de la Société Française de Microbiologie, Société Française de Microbiologie, Recommendations 2010, p 44.
Antimicrobial
Agent/ClassDisk
ContentZone Diameter,
nearest whole mmMIC Interpretive
Standard (µg/ml)S R S R PENICILLINS Penicillin G1
Amoxicillin1
Oxacillin1- 2
-
5 µg-
-
≥ 18-
-
-≤ 0.06
≤ 0.12
-> 0.25
> 1
-CEPHEMS Cefotaxime2
Ceftriaxone230 µg
30 µg-
--
-≤ 0.12
≤ 0.12-
-PHENICOLS Chloramphenicol 30 µg ≥ 30 - ≤ 2 > 4 ANSAMYCINS Rifampicin3 30 µg ≥ 30 - ≤ 0.25 - FLUOROQUINOLONES Ciprofloxacin - - - ≤ 0.03 > 0.06
Footnotes
1Disk diffusion tests with amoxicillin and penicillin G for N. meningitidis are unreliable. Testing can be performed using 5 µg/ml oxacillin disks. Diameters of ≥ 18 mm are sensitive to penicillin G, but the MICs against penicillin G and/or amoxicillin should be determined for those diameters of < 18 mm.
2- = no data available.
3Used only for prophylaxis.
Table 8. MIC interpretive standards for N. meningitidis as recommended by EUCAST
Data is from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) website. http://www.eucast.org, version 1.3, December 2010.
EUCAST does not yet have interpretive guidelines for Kirby-Bauer disk diffusion for N. meningitidis.
Antimicrobial
Agent/ClassMIC (µg/ml)
Interpretive StandardS R PENICILLINS Benzylpenicillin
Ampicillin≤ 0.064
≤ 0.125> 0.25
> 1.0CEPHEMS Cefotaxime1
Ceftriaxone1≤ 0.125
≤ 0.125> 0.125
> 0.125CARBAPENEMS Meropenem1,2 ≤ 0.25 > 0.25 ANSAMYCINS Rifampicin ≤ 0.25 > 0.25 TETRACYCLINES Tetracycline3 ≤ 1.0 > 2.0 FLUOROQUINOLONES Ciprofloxacin4 ≤ 0.032 > 0.064 PHENICOLS Chloramphenicol ≤ 2.0 > 4.0
Footnotes
1Strains with MIC values above the susceptible breakpoint are very rare or not yet reported. The identification and antimicrobial susceptibility tests on any such isolate must be repeated and if the result is confirmed, the isolate should be sent to a reference laboratory. Until there is evidence regarding clinical response for confirmed isolates with MIC above the current resistant breakpoint they should be reported resistant.
2These breakpoints relate to meningitis only.
3Tetracycline can be used to determine minocycline susceptibility for prophylaxis of N. meningitidis infections.
4Breakpoints apply only to use in the prophylaxis of meningococcal disease.
-
-
Minimal inhibitory concentration testing of N. meningitidis by antimicrobial gradient strips
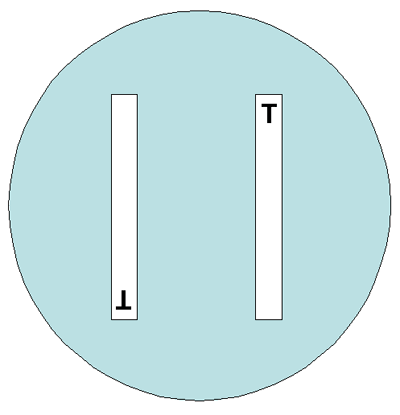
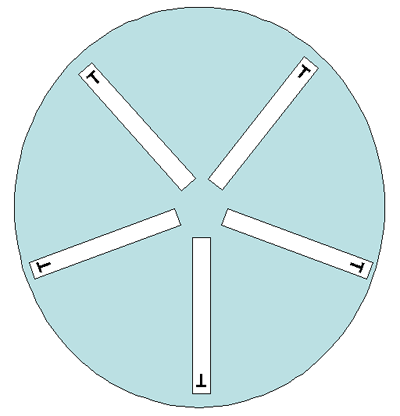
Either 150-mm or 100-mm plates can be used for the gradient strip testing, depending on the number of antimicrobial agents to be tested per isolate. Two different gradient strips can be placed in opposite gradient directions on a 100-mm plate (Figure 4). Although one manufacturer states that up to six gradient strips can be used on a 150-mm plate, this laboratory manual suggests that in order to avoid overlapping zones of inhibition of growth, not more than five gradient strips be used on a 150-mm plate, with the end with the lowest concentration of antibiotic placed towards the center of the plate (Figure 5). Depending on the bacteria/antimicrobial combinations, five strips on a 150-mm plate may lead to overlapping ellipses. If this occurs, testing should be repeated using four strips per plate.
Isolates to be tested should be subcultured onto a chocolate agar plate and incubated in a CO2-enhanced atmosphere (5% CO2 in a CO2-incubator or candle-extinction jar) at 35±2°C for 20-24 hours prior to testing. If the organism has been frozen, it should be subcultured twice when it is removed from the freezer before proceeding with susceptibility testing.
Mueller-Hinton agar with 5% sheep blood is used when testing N. meningitidis isolates with gradient strips. Warm the cation-adjusted Mueller-Hinton broth (CAMHB) to 35°C before using. Allow the gradient strips that will be used in the batch of testing to warm to room temperature (25°C). It is recommended to follow the directions on the package insert included withthe gradient strips.
-
Using a sterile cotton-tip applicator, touch the surface of one to four morphologically identical, isolated colonies. Immerse the applicator into a tube containing sterile CAMHB. Rub the applicator against the wall of the tube slightly to release a small amount of growth into the liquid. Cap the tube and mix the cells using a vortex to form a suspension, being careful not to form froth or bubbles in the suspension when mixing the cells. Adjust the turbidity of the inoculum to that of a 0.5 McFarland turbidity standard (approximately 1 to 4 x108 CFU/ml). Preparation of aMcFarland turbidity standard is described inthe Annex. If the turbidity of the inoculum is greater than the standard, dilute it with CAMHB to equal the turbidity of the standard. This suspension must be used within 15 minutes.
- Perform regular colony counts to verify that the density of the inoculum suspension is correct. For example, dilute the suspension 1:100 and subculture 10 µl onto the recommended media. An acceptable inoculum should give approximately 100-500 colonies. It is not necessary to perform colony counts on every isolate tested.
- Immerse a sterile cotton-tipped swab into the adjusted inoculum. Remove excess liquid by pressing the swab tip against the inside of the tube. Inoculate the entire surface of a 15x150-mm Mueller-Hinton agar with 5% sheep blood plate three times with the same swab of inoculum, rotating the plate 60 degrees after each inoculation to ensure even distribution of the inoculum and confluent growth of the bacteria. Use a single swab of inoculum and do not return the swab to the broth after each rotation.
- Allow the inoculum to dry on the surface of the plate (which should take approximately 5-10 minutes). Be sure the plate is entirely dry before proceeding.
- When the surface of the inoculated plate is dry and the gradient strips are at room temperature, place the antimicrobial gradient strips onto the agar with an applicator or sterile forceps. Make sure that the printed MIC values are facing upward (i.e., that the bottom surface of the strip containing the antimicrobial gradient is in contact with the agar). Alternatively, robotic gradient strip applicators are available from some manufacturers. Once applied, it is important to not move the antimicrobial gradient strips as the antibiotic diffuses into the agar immediately upon contact.
- Return the antimicrobial gradient strips that will not be used in this batch of testing to the -20°C freezer (some strips can be stored at 4°C. Follow the manufacturer’s instructions).
- Incubate the plates in an inverted position in a 5% CO2 atmosphere for 18–22 hours at 37°C. A candle-extinction jar may be used if a CO2-incubator is not available. Because N. meningitidis grows well in a humid atmosphere, laboratorians may choose to add a shallow pan of sterile water to the bottom of the incubator or add a dampened paper towel to the candle-extinction jar.
- After incubation, an ellipse of bacterial growth will have formed on the plate around the strip and the test can be read (see below). QC results must be reviewed before reading and interpreting the MIC.
Reading and interpreting the gradient strips
Read the MIC at the point where the zone of inhibition intersects the MIC scale on the strip. Use oblique light to carefully examine the end point. A magnifying glass may be used if needed. Read at the point of complete inhibition of all growth including hazes. A reading guide for the gradient strips, which shows organism related effects, drug-related effects, resistance mechanism-related effects, and technical and handling effects can be found at: http://www.biomerieux-diagnostics.com/sites/clinic/files/etest-reading-guide-aerobic-bacteria.pdf [1 page].
Record the QC results first. If zones produced by the QC strain are out of the expected ranges (see Tables 2, 3, and 5, depending on the strains and guidelines being used), the laboratorian should consider possible sources of error. Because antimicrobial susceptibility test results can be affected by many factors not necessarily associated with the actual susceptibility of the organism (e.g., inoculum size, growth phase, agar depth, storage, time, and others), QC practices must be followed carefully (see Section II above). If all antimicrobial agents are in the control range, read the test MICs. Note any unusual observations such as a zone of incomplete killing (trailing endpoints) or single colonies growing within the ellipse.
The gradation marks on the gradient strip correspond to the standard 2-fold dilution concentrations for the agar dilution method, but also include increments between those standard values. The standard values are used for interpretation and reporting of antimicrobial susceptibility test results. It is advised that both the actual reading of the value from the strip and the next-higher standard value (i.e., the value to be used for interpretation) be included in the laboratory records for testing of the strain. For example, if testing susceptibility of an isolate to penicillin, an MIC recorded from the gradations on the gradient strip might be 0.094 µg/ml; however, the reported MIC would be 0.125 µg/ml.
The manufacturer of the gradient strips recommends following the MIC breakpoints developed for agar and broth microdilution. The interpretive standards and MIC breakpoints recommend by CLSI (Table 6), CA-SFM (Table 7), and EUCAST (Table 8) are given.
-
-
- Antimicrobial susceptibility testing of H. influenzae
This laboratory manual describes susceptibility testing of H. influenzae by the disk diffusion method and by the antibiotic gradient strip testing method. Although disk diffusion will provide information as to whether a strain is susceptible, intermediate, or resistant, the gradient strip method provides more detailed information about the minimal inhibitory concentration (MIC) of an antimicrobial agent. In addition, testing for strains employing a β-lactamasewill be described.
The β-lactamase usually observed circulating in H. influenzae is a TEM-type β-lactamase, which inactivates some antibiotics belonging to the b-lactam family such as penicillin, ampicillin, and amoxicillin, but, fortunately, does not inactivate third generation cephalosporins such as ceftriaxone or cefotaxime. This β-lactamaseis inhibited by clavulanic acid. However, as clavulanic acid does not cross the blood-brain barrier well, the association of amoxicillin and clavulanic acid must not be employed for the treatment of a meningitis due to H. influenzae. In this case, ceftriaxone or cefotaxime have to be recommended. Resistance due to a reduction of affinity to the Penicillin Binding Protein (PBP) is rare but has been reported (3).
-
Testing H. influenzae for β-lactamase production
A rapid β-lactamasetest may yield clinically relevant information earlier than the results of antimicrobial susceptibility testing, so it should be performed as soon as a H. influenzae is identified.
-
Nitrocefin-based tests are the preferred method. The reagent is composed of paper disks impregnated with chromogenic cephalosporin, which releases a red compound on hydrolysis by a β-lactamase.
Performing the test: The disks can be stored in their cartridge at 2-8°C until the expiration date. After opening the cartridge, disks can be stored for 2 months at 2-8°C.
- Allow the tube containing the cartridge to come to room temperature (25°C).
- Moisten a disk with sterile distilled water.
- Collect a few isolated colonies of the strains to be tested and spread them over the surface of the disk.
Reading and interpretation
The appearance of a red color reveals a positive reaction.
The reaction is negative if no color has appeared after 30 minutes.Quality control strains
Staphylococcus aureus ATCC 29213 result +
Enterococcus faecalis ATCC 29212 result -
-
-
Antimicrobial susceptibility testing of H. influenzae by Kirby-Bauer disk diffusion
The antibiotic susceptibility testing of H. influenzaeusing the disk diffusion method will provide information as to whether a strain is susceptible, intermediate, or resistant to an antimicrobial. Dilution methods or antimicrobial gradient strips can be used to accurately determine the MICs to antimicrobials, but are not necessarily better than disk diffusion in providing reliable information about determining whether the isolate is susceptible, intermediate, or resistant to an antimicrobial. Laboratories must use standardized procedures to guarantee the accuracy and reproducibility of antibiotic susceptibility testing.
Media and disks for antimicrobial susceptibility testing
For H. influenzae, antimicrobial susceptibility can be determined using the disk diffusion method. The method presented in this chapter is a modification of the Kirby-Bauer technique that has been standardized by CLSI. If performed precisely according to the following protocol, this method will provide data that can reliably predict the in vivo effectiveness of the drug in question. The accuracy and reproducibility of this test is dependent on the consistent use of a standard set of procedures in the laboratory.
The optimal medium is Haemophilus test medium (HTM). The Mueller-Hinton agar used to make HTM should be thymidine free to obtain consistent results if susceptibility to cotrimoxazole is to be tested. HTM medium consists of the following ingredients: Mueller-Hinton agar supplemented with 15 µg/ml NAD, 15 µg/ml bovine hemin, and 5 mg/ml yeast extract. The pH is adjusted to 7.2 to 7.4. Recommended agents tested are ampicillin, ceftriaxone and/or cefotaxime, and chloramphenicol, which are antibiotics commonly used for the treatment of meningitis.
The 10 µg-ampicillin disk predicts both intrinsic (PBP-mediated) and β-lactamase mediated penicillin and ampicillin resistance of H. influenzae. A 30 µg-chloramphenicol disk is used for predicting resistance to chloramphenicol, and a 30 µg-ceftriaxone and/or cefotaxime disk is used for predicting susceptibility to these antibiotics. The zone diameter sizes can only be properly interpreted when HTM is used. The results have to be compared to standards that have been validated, such as those recommended by CLSI (Table 9), CA-SFM (Table 10), and EUCAST (Table 11).
Quality control
QC tests should be performed once per week if susceptibility tests are performed daily or with every group of tests when testing is done less frequently than every day. QC tests have to be done for each new batch of test medium or new lot of disks. If the results found for the control strain are accurate, the procedure is assumed to be correct. If this is not the case, the tests can be affected by variation in media, inoculum size, incubation time, temperature, the depth of the agar in the plate (uniformly 3-4 mm), the pH (between 7.2-7.4), disk potency, the purity of the culture for inoculum, or if the concentration of bacteria does not approximate the 0.5 McFarland turbidity standard. CLSI, CA-SFM, and EUCAST list recommended QC strains and test limits (Tables 1, 3, and 4). A laboratory should choose which QC strain(s) to use based on the antimicrobials to be tested for susceptibility.
Table 9. Kirby-Bauer disk diffusion zone diameter interpretive standards and MIC interpretive standards for H. influenzae as recommended by CLSI
Copyrighted material used with permission from the Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, PA, USA 19087, www.clsi.org.
CLSI document M100-S21; 2011, pp 88-91.
Antimicrobial
Agent/ClassDisk
ContentZone Diameter,
Nearest Whole mmMIC Interpretive
Standard (µg/ml)S I R S I R PENICILLINS Ampicillin1 10 µg ≥ 22 19-21 ≤ 18 ≤ 1 2 ≥ 4 CEPHEMS Cefotaxime2
Ceftriaxone230 µg
30 µg≥ 26
≥ 26-
--
-≤ 2
≤ 2-
--
-PHENICOLS Chloramphenicol 30 µg ≥ 29 26-28 ≤ 25 ≤ 2 4 ≥ 8
Footnotes
1In most cases, a direct b-lactamase test can provide a rapid means of detecting ampicillin and amoxicillin resistance. The majority of isolates of H. influenzae that are resistant to ampicillin and amoxicillin produce a TEM-type β-lactamase.
2For some antimicrobial agents, the absence or rare occurrence of resistant strains precludes defining any results categories other than “susceptible”.
Table 10. Kirby-Bauer disk diffusion zone diameter and MIC interpretive standards for H. influenzae as recommended by CA-SFM
Comite de L’Antibiogramme de la Société Française de Microbiologie, Société Française de Microbiologie, Recommendations 2010, pp 42-43.
Antimicrobial
Agent/ClassDisk
ContentZone Diameter,
Nearest Whole mmMIC (µg/ml)
Interpretive StandardS R S R PENICILLINS Ampicillin1,2 2 µg ≥ 20 < 20 ≤ 1 >1 CEPHEMS Ceftriaxone or Cefotaxime3 - - - ≤ 0.12 - PHENICOLS Chloramphenicol 30 µg ≥ 30 < 26 ≤ 1 > 2
Footnotes
1A positive direct chromogenic β-lactamase test predicts resistance to penicillin, ampicillin, and amoxicillin.
2β-lactamase-negative, ampicillin-resistant (BLNAR) strains are rare, but detection of decreased susceptibility to beta-lactams in BLNAR strains is possible using a 2 µg ampicillin disk (diameter < 20 mm) or a 30 µg cephalothin disk (diameter < 17 mm).
3Neither clinical failure nor resistance has been reported for these antimicrobial agents, thus the criteria for interpretative breakpoints has not been established for any category other than susceptible.
Table 11. Kirby-Bauer Disk Diffusion Zone Diameter and MIC Interpretive Standards for H. influenzae as Recommended by EUCAST.
Data from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) website. http://www.eucast.org. Version 1.3, December, 2010.
Antimicrobial
Agent/ClassDisk
ContentZone Diameter,
Nearest Whole mmMIC (µg/mL)
Interpretive StandardS R S R PENICILLINS Ampicillin1,2 2 µg ≥ 16 < 16 ≤ 1 >1 CEPHEMS Cefotaxime3
Ceftriaxone35 µg
30 µg≥ 22
≥ 27< 22
< 27≤ 0.125
≤ 0.125-
-PHENICOLS Chloramphenicol 30 µg ≥ 30 < 26 ≤ 1 > 2
Footnotes
1Report β-lactamase positive strains resistant to penicillins without β-lactamase inhibitors.
2Breakpoints relate only to β-lactamase negative strains. Strains may be resistant to penicillins, aminopenicillins, cephalosporins and/or carbapenems due to changes in penicillin binding proteins (BLNAR, β-lactamase negative ampicillin resistant) and a few strains have both resistance mechanisms (BLPACR, β-lactamase positive, amoxicillin/clavulanate resistant).
3Neither clinical failure nor resistance has been reported for these antimicrobial agents, thus the criteria for interpretative breakpoints has not been established for any category other than susceptible.
Antimicrobial susceptibility testing procedure of H. influenzae by Kirby-Bauer disk diffusion
Either 150-mm or 100-mm plates can be used for Kirby-Bauer disk diffusion depending on the number of antimicrobial agents to be tested per isolate. CLSI guidelines state that no more than two disks can be used on a 100-mm plate and up to five disks can be used on a 150-mm plate (Figure 1).
Isolates to be tested should be subcultured onto a supplemented chocolate agar plate and incubated in a CO2-enhanced atmosphere (5% CO2 in a CO2-incubator or candle-extinction jar) at 35±2°C for 20-24 hours prior to testing. If the organism has been frozen, it should be subcultured twice when it is removed from the freezer before proceeding with susceptibility testing.
Remove agar plates from the refrigerator and allow them to come to room temperature (25°C) before inoculating. If HTM broth is to be used to make the 0.5 McFarland, warm it to 35°C before using. Allow the antibiotic disks that will be used in the batch of testing to warm to room temperature (25°C).
- Remove agar plates from the refrigerator and allow them to come to room temperature (25°C) before inoculating. If HTM broth is to be used to make the 0.5 McFarland, warm it to 35°C before using. Allow the antibiotic disks that will be used in the batch of testing to warm to room temperature (25°C).
- Perform regular colony counts to verify that the density of the inoculum suspension is correct. For example, dilute the suspension 1:100 and subculture 10 µl onto the recommended media. An acceptable inoculum should give approximately 100-500 colonies. It is not necessary to perform colony counts on every isolate tested.
- Immerse a sterile cotton-tipped swab into the adjusted inoculum. Remove excess liquid by pressing the swab tip against the inside of the tube. Inoculate the entire surface of a HTM plate three times with the same swab of inoculum, rotating the plate 60 degrees after each inoculation to ensure even distribution of the inoculum and confluent growth of the bacteria. Use a single swab of inoculum and do not return the swab to the broth after each rotation.
- Allow the inoculum to dry on the surface of the plate (which should take approximately 5-10 minutes). Be sure the plate is entirely dry before proceeding, but do not exceed 15 minutes.
- When the surface of the inoculated plate is dry and the disks are at room temperature, place the disks onto the agar with an applicator or sterile forceps. Make sure that the disks are spaced enough distance apart on the agar so the zones of inhibition do not overlap (Figure 1). Press down on the disks to ensure complete contact with the agar surface. Alternatively, a mechanical disk dispenser can be used. Once applied, it is important to not move the antibiotic disks as the antibiotic will begin to diffuse immediately upon contact with the plate.
- Incubate the plates in an inverted position in a 5% CO2 atmosphere or candle jar for 20–24 hours at 35±2°C.
- After overnight incubation, measure the diameter of each zone of inhibition with a ruler or calipers (Figure 2). Measurements should be performed in a biosafety cabinet, if possible. The zones of inhibition on the media are measured by holding the Petri dish a few inches above a black, nonreflective background illuminated with reflected light. Use either calipers or a ruler with a handle attached for these measurements, holding the ruler over the center of the surface of the disk when measuring the inhibition zone (Figures 1 and 2).
- Care should be taken not to touch the disk or surface of the agar. Decontaminate the ruler occasionally to prevent transmission of the bacteria. In all measurements, the zones of inhibition are measured as the diameter from the edges of the last visible colony of the unaided eye. Record the results to the nearest millimeter (mm).
- Interpret the antimicrobial susceptibility of the strain being tested (and check that results for the QC strains are within the acceptable control range) by comparing the results to the CLSI (Table 9), CA-SFM (Table 10), or EUCAST (Table 11) standard zone sizes. See Figure 3 for a sample worksheet for recording antimicrobial susceptibility test results for H. influenzae.
- Remove agar plates from the refrigerator and allow them to come to room temperature (25°C) before inoculating. If HTM broth is to be used to make the 0.5 McFarland, warm it to 35°C before using. Allow the antibiotic disks that will be used in the batch of testing to warm to room temperature (25°C).
-
Minimal inhibitory concentration testing of H. influenzae by antimicrobial gradient strips
In reference laboratories, it is necessary to precisely monitor for any changes in the MICs of the isolates. It can be done either by an antibiotic dilution method or using antimicrobial gradient strips. The gradient strip method is convenient and reliable, giving 85-100% correlation with the dilution method. Gradient strips must be stored at -20°C or 4°C (follow the manufacturer’s recommendations). Either 150-mm or 100-mm plates can be used for the gradient strip testing, depending on the number of antimicrobial agents to be tested per isolate. Two different gradient strips can be placed in opposite gradient directions on a 100-mm plate (Figure 4). Although one manufacturer states that up to six gradient strips can be used on a 150-mm plate, this laboratory manual suggests that in order to avoid overlapping zones of inhibition of growth, not more than five gradient strips be used on a 150-mm plate, with the end with the lowest concentration of antibiotic placed towards the center of the plate (Figure 5). Depending on the bacteria/antimicrobial combinations, five strips on a 150 mm plate may lead to overlapping ellipses. If this occurs, testing should be repeated using four strips per plate.
Isolates to be tested should be subcultured onto a supplemented chocolate agar plate and incubated in a CO2-enhanced atmosphere (5% CO2 in a CO2-incubator or candle-extinction jar) at 35±2°C for 20-24 hours prior to testing. If the organism has been frozen, it should be subcultured twice when it is removed from the freezer before proceeding with susceptibility testing.
Remove agar plates from the refrigerator and allow them to come to room temperature (25°C) before inoculating. If HTM broth is to be used to make the 0.5 McFarland, warm it to 35°C before using. Allow the gradient strips that will be used in the batch of testing to warm to room temperature (25°C). It is recommended to follow the directions on the package insert included withthe gradient strips
- Using a sterile cotton-tip applicator, touch the surface of one to four morphologically identical, isolated colonies. Immerse the applicator into a tube containing sterile HTM broth or saline. Rub the applicator against the wall of the tube slightly to release a small amount of growth into the liquid. Cap the tube and mix the cells using a vortex to form a suspension, being careful not to form froth or bubbles in the suspension when mixing the cells. Adjust the turbidity of the inoculum to that of a 0.5 McFarland turbidity standard (approximately 1 to 4 x 108 CFU/ml). Preparation of aMcFarland turbidity standard is described inthe Annex. If the turbidity of the inoculum is greater than the standard, dilute it with HTM broth or saline to equal the turbidity of the standard. This suspension must be used within 15 minutes.
- Perform regular colony counts to verify that the density of the inoculum suspension is correct. For example, dilute the suspension 1:100 and subculture 10 µl onto the recommended media. An acceptable inoculum should give approximately 100-500 colonies. It is not necessary to perform colony counts on every isolate tested.
- Immerse a sterile cotton-tipped swab into the adjusted inoculum. Remove excess liquid by pressing the swab tip against the inside of the tube. Inoculate the entire surface of a HTM plate three times with the same swab of inoculum, rotating the plate 60 degrees after each inoculation to ensure even distribution of the inoculum and confluent growth of the bacteria. Use a single swab of inoculum and do not return the swab to the broth after each rotation.
- Allow the inoculum to dry on the surface of the plate (which should take approximately 5-10 minutes). Be sure the plate is entirely dry before proceeding, but do not exceed 15 minutes.
- When the surface of the inoculated plate is dry and the gradient strips are at room temperature, place the antimicrobial gradient strips onto the agar with an applicator or sterile forceps. Make sure that the printed MIC values are facing upward (i.e., that the bottom surface of the strip containing the antimicrobial gradient is in contact with the agar). Alternatively, robotic gradient strip applicators are available from some manufacturers. Once applied, it is important to not move the antimicrobial gradient strips as the antibiotic diffuses into the agar immediately upon contact.
- Return the antimicrobial gradient strips that will not be used in this batch of testing to the -20°C freezer (some strips can be stored at 4°C. Follow the manufacturer’s instructions).
- Incubate the plates in an inverted position in a 5% CO2 atmosphere or candle jar for 20–24 hours at 35±2°C.
- After incubation, an ellipse of bacterial growth will have formed on the plate around the strip and the test can be read (see below). QC results must be reviewed before reading and interpreting the MIC.
Reading and interpreting the gradient strips
Read the MIC at the point where the zone of inhibition intersects the MIC scale on the strip. Use oblique light to carefully examine the end point. A magnifying glass may be used if needed. Read at the point of complete inhibition of all growth including hazes. A reading guide for the gradient strips, which shows organism related effects, drug-related effects, resistance mechanism-related effects, and technical and handling effects can be found at: http://www.abbiodisk.com/pdf/pi/75002206.pdf [1.68 MB, 2 pages].
Record the QC results first. If zones produced by the control strain are out of the expected ranges (see Tables 2, 3, and 5, depending on the strains and guidelines being used), the laboratorian should consider possible sources of error. Because antimicrobial susceptibility test results can be affected by many factors not necessarily associated with the actual susceptibility of the organism (e.g., inoculum size, growth phase, agar depth, storage, time, and others), QC practices must be followed carefully (see Section II above). If all antimicrobial agents are in the control range, read the test MICs. Note any unusual observations such as a zone of incomplete killing (trailing endpoints) or single colonies growing within the ellipse.
The gradation marks on the gradient strip correspond to the standard 2-fold dilution concentrations for the agar dilution method, but also include increments between those standard values. The standard values are used for interpretation and reporting of antimicrobial susceptibility test results. It is advised that both the actual reading of the value from the strip and the next-higher standard value (i.e., the value to be used for interpretation) be included in the laboratory records for testing of the strain. For example, if testing susceptibility of an isolate to penicillin, an MIC recorded from the gradations on the gradient strip might be 0.094 µg/ml; however, the reported MIC would be 0.125 µg/ml.
The manufacturer of the gradient strips recommends following the MIC breakpoints developed for agar and broth microdilution. The interpretive standards and MIC breakpoints recommend by CLSI (Table 9), CA-SFM (Table 10), and EUCAST (Table 11) are given.
- Using a sterile cotton-tip applicator, touch the surface of one to four morphologically identical, isolated colonies. Immerse the applicator into a tube containing sterile HTM broth or saline. Rub the applicator against the wall of the tube slightly to release a small amount of growth into the liquid. Cap the tube and mix the cells using a vortex to form a suspension, being careful not to form froth or bubbles in the suspension when mixing the cells. Adjust the turbidity of the inoculum to that of a 0.5 McFarland turbidity standard (approximately 1 to 4 x 108 CFU/ml). Preparation of aMcFarland turbidity standard is described inthe Annex. If the turbidity of the inoculum is greater than the standard, dilute it with HTM broth or saline to equal the turbidity of the standard. This suspension must be used within 15 minutes.
-
- Antimicrobial susceptibility testing of S. pneumoniae
This section describes the optimal media, inoculum, antimicrobial agents to test, incubation conditions, and interpretation of results for S. pneumoniae by the disk diffusion method and the antimicrobial gradient strip method. Although disk diffusion will provide information for most antimicrobial agents regarding interpretation of a strain as susceptible, intermediate, or resistant, the antimicrobial gradient strip test provides general information about the MIC of antibiotic. The accuracy and reproducibility of this test are dependent on following a standard set of procedures and conditions in laboratories on an everyday basis.
Quality control
Quality control tests must be performed as part of the normal laboratory routine. To verify that antimicrobial susceptibility test results are accurate, at least one control organism should be included with each test or new set of testing conditions. CLSI, CA-SFM, and EUCAST list recommended QC strains and test limits (Table 1-5). A laboratory should choose which QC strain(s) to use based on the antimicrobials to be tested for susceptibility. Further information about trouble-shooting out of range quality control results can be found in Section II above.
-
Antimicrobial susceptibility testing of S. pneumoniae by Kirby-Bauer disk diffusion
Mueller-Hinton agar medium supplemented with 5% sheep blood is recommended for determining the antimicrobial susceptibility of S. pneumoniae specimens by disk diffusion. The agar plates should have a uniform depth of 3-4 mm. Prepare the inoculum for antimicrobial susceptibility testing of S. pneumoniae from fresh pure cultures of S. pneumoniae (grown overnight on blood or chocolateagar). Prepare cell suspensions of the bacteria to be tested in sterile physiologicalsaline or Mueller-Hinton broth. A cell suspension equal to a density of a 0.5 McFarland turbidity standard is used for the inoculum (approximately 1 to 4 x 108 CFU/ml). Preparation of aMcFarland turbidity standard is described in theAnnex.
- Suspend viable colonies from an overnight sheep blood or chocolate agar plate in a tube of broth or saline to achieve a bacterial suspension equivalent to a 0.5 McFarland turbidity standard; be careful not to form froth or bubbles in the suspension when mixing the cells with the broth.
-
Compare the density of the suspension to the 0.5 McFarland turbidity standard by holding the suspension and McFarland turbidity standard in front of a light against a white background with contrasting black lines. If the density is too heavy, the suspension should be diluted with saline or broth (whichever was used to make the suspension). If the density is not sufficient, additional bacteria should be added to the suspension. This suspension should be used within 15 minutes.
- Perform regular colony counts to verify that the density of the inoculum suspension is correct. For example, dilute the suspension 1:100 and subculture 10 µl onto the recommended media. An acceptable inoculum should give approximately 100-500 colonies. It is not necessary to perform colony counts on every isolate tested.
- When the proper density is achieved, dip a cotton swab into the bacterial suspension. Lift it out of the broth and remove excess fluid by pressing and rotating the swab against the wall of the tube.
- Use the swab to inoculate the entire surface of the supplemented Mueller-Hinton agar plate three times, rotating the plate 60 degrees between each inoculation. Use the same swab with each rotated streak, but do not re-dip the swab in the inoculum (i.e., the bacterial cell suspension).
- Allow the inoculum to dry before placing the disks on the plates. Drying usually takes only a few minutes and should take no longer than 15 minutes. If drying takes longer than 15 minutes, use a smaller volume of inoculum in the future by pressing more liquid out of the swab.
- After the plate is dry, place the antimicrobial disks on the plates (Figure 1). Use sterile forceps to place the disks on the Mueller-Hinton agar and tap them gently to ensure they adhere to the agar. Alternatively, a mechanical disk dispenser can be used. Diffusion of the drug in the disk begins immediately; therefore, once a disk contacts the agar surface, the disk should not be moved.
- Incubate the plates in an inverted position in a 5% CO2 atmosphere for 20-24 hours at 37°C. A candle-extinction jar may be used if a CO2-incubator is not available.
- If this is a new batch of Mueller-Hinton agar, the antimicrobial disks are new, or it is an otherwise appropriate time to perform QC, follow steps 1 through 7 above and run parallel tests on the reference strain(s). Appropriate disk diffusion zone sizes for the reference QC strains are included in Tables 1, 3 and 4.
- After overnight incubation, measure the diameter of each zone of inhibition with a ruler or calipers. The zones of inhibition are measured from the top surface of the plate with the top removed. Use either calipers or a ruler with a handle attached for these measurements, holding the ruler over the center of the surface of the disk when measuring the inhibition zone (Figure 1).
- Care should be taken not to touch the disk or surface of the agar. Decontaminate the ruler occasionally to prevent transmission of the bacteria. In all measurements, the zones of inhibition are measured as the diameter from the edges of the last visible colony. Record the results in millimeters (mm). Figure 3 provides a sample form for recording results.
- Interpret the antimicrobial susceptibility of the strain being tested (and check that results for the QC strain(s) are within the acceptable control range) by comparing the results to the CLSI (Table 12), CA-SFM (Table 13), or EUCAST (Table 14) standard zone sizes.
-
Minimal inhibitory concentration testing of S. pneumoniae by antimicrobial gradient strips
For S. pneumoniae, disk diffusion testing indicates whether an organism is susceptible or resistant to an antimicrobial for most agents. However, disk diffusion testing for pneumococcal isolates using oxacillin (a penicillin family antibiotic) is not sufficient to distinguish between complete and intermediate resistance. For surveillance purposes, a laboratory may want to quantify the results of the oxacillin disk diffusion test by performing MIC testing of penicillin or any other b-lactam antibiotic that would be used for treatment. As mentioned earlier in this manual, MIC testing by dilution can be expensive and challenging, and because of the technical complexity required for these tests, countries that do not currently do MIC testing by dilution should utilize an international reference laboratory rather than developing the assay in-country. In countries where MIC testing is done at more than one laboratory,
Table 12. Kirby-Bauer disk diffusion zone diameter interpretive standards and MIC interpretive standards for S. pneumoniae as recommended by CLSI
Copyrighted material used with permission from the Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, PA, USA 19087, www.clsi.org.
CLSI document M100-S21; 2011, pp 96-99.
Antimicrobial
Agent/ClassDisk
ContentZone Diameter,
Nearest Whole mmMIC Interpretive
Standard (µg/ml)S I R S I R PENICILLINS Penicillin1,2,3
parenteral (meningitis)1 µg4 oxacillin ≥ 20 - - ≤ 0.06 - ≥ 0.12 CEPHEMS Cefotaxime2,3,5,6
(meningitis)- - - - ≤ 0.5 1 ≥ 2 MACROLIDES Erythromycin6 15 µg ≥ 21 16-20 ≤ 15 ≤ 0.25 0.5 ≥ 1 FOLATE PATHWAY INHIBITORS8,9 Trimethoprim-sulfamethoxazole
(co-trimoxazole)1.25/
23.75µg≥ 19 16-18 ≤ 15 ≤ 0.5/9.5 1/19-2/38 ≥ 4/76 TETRACYCLINES Tetracycline10 30 µg ≥ 23 19-22 ≤ 18 ≤ 2 4 ≥ 8 FLUOROQUINOLONES Levofloxacin 5 µg ≥ 17 14-16 ≤ 13 ≤ 2 4 ≥ 8 LINCOSAMIDES Clindamycin 2 µg ≥ 19 16-18 ≤ 15 ≤ 0.25 0.5 ≥ 1 KETOLIDES Telithromycin 15 µg ≥ 19 16-18 ≤ 15 ≤ 1 2 ≥ 4
Footnotes
1Rx: Use of penicillin in meningitis requires therapy with maximum doses of intravenous penicillin (e.g., at least 3 million units every four hours in adults with normal renal function).
2Penicillin, cefotaxime, and ceftriaxone should be tested by a reliable MIC method and reported routinely with CSF isolates of S. pneumoniae.
3For CSF isolates, report only meningitis interpretations.
4Isolates of pneumococci with oxacillin zone sizes of ≥ 20 mm are susceptible (MIC ≤ 0.06 mg/ml) to penicillin. Penicillin and cefotaxime, ceftriaxone, or meropenem MICs should be determined for those isolates with oxacillin zone diameters of ≤ 19 mm, because zones of ≤ 19 mm occur with penicillin-resistant, intermediate, or certain susceptible strains. For isolates with oxacillin zones ≤ 19 mm, do not report penicillin as resistant without performing a penicillin MIC test.
5Rx: Use of cefotaxime or ceftriaxone in meningitis requires therapy with maximum doses.
6Breakpoints for isolates from cases of meningitis are identical to ceftriaxone.
7Susceptibility and resistance to azithromycin, clarithromycin, and dirithromycin can be predicted by using erythromycin.
8The Mueller-Hinton agar used for this test should be thymidine free to obtain accurate results.
9Trimethoprim-sulfamethoxazole is a combination of two drugs used in treatment (cotrimoxazole). The 1:20 ratio is that at which the greatest synergy in treatment has been demonstrated in serum. The disks are impregnated with 1.25 µg trimethoprim and 23.75 µg sulfamethoxazole. The MIC breakpoints mimic the 1/20 ratio.
10Organisms susceptible to tetracycline are also considered susceptible to doxycycline and minocycline.
Table 13. Kirby-Bauer disk diffusion zone diameter and MIC interpretive standards for S. pneumoniae as recommended by CA-SFM
Comite de L’Antibiogramme de la Société Française de Microbiologie, Société Française de Microbiologie, Recommendations 2010, pp 38-39.
Antimicrobial
Agent/ClassDisk
ContentZone Diameter,
Nearest Whole mmMIC Interpretive
Standard (µg/ml)S R S R PENICILLINS Penicillin G1,2,3 5 µg Oxacillin - - ≤ 0.06 > 2 CEPHEMS Cefotaxime1,2,3
Ceftriaxone1,2,3-
--
-≤ 0.5
≤ 0.5> 2
> 2MACROLIDES Erythromycin4 15 IU ≥ 26 < 24 ≤ 0.25 > 0.5 FOLATE PATHWAY INHIBITORS5,6,7 Trimethoprim-sulfamethoxazole 1.25/23.75 µg ≥ 16 < 10 ≤ 2/38 >8/152 TETRACYCLINES Tetracycline8 30 IU ≥ 23 < 21 ≤ 1 > 2 FLUOROQUINOLONES Levofloxacin9 5 µg ≥ 17 < 17 ≤ 2 > 2 LINCOSAMIDES Clindamycin 15 µg ≥ 21 < 17 ≤ 0.5 > 0.5 KETOLIDES Telithromycin10 15 µg ≥ 24 < 21 ≤ 0.25 > 0.5
Footnotes
1Disk diffusion tests with penicillin G for S. pneumoniae are unreliable, but testing can be performed using 5 µg/ml oxacillin (OXA-5) disks according to the following criteria: Diameters of ≥ 26 mm using OXA-5 disks are sensitive to penicillin G and other β-lactams. Diameters of < 26 mm using OXA-5 disks have reduced susceptibility.
2This test cannot assess the level of resistance to penicillin G or other β-lactams. Using disks of other β-lactam antibiotics cannot determine the level of resistance to these β-lactams.
3In cases of severe infection, clinical failure, or any strain with reduced susceptibility, it is necessary to determine the MIC to penicillin G and at least one of β-lactam with pharmacodynamic properties which are consistent with therapeutic efficacy (amoxicillin, cefotaxime, or ceftriaxone). Strains categorized as intermediate (MIC of > 0.064 µg/ml to >2 µg/ml) or even with low level resistance should be considered resistant in the case of meningitis, but sensitive to high doses in the case of respiratory infections.
4Interpretation valid for azithromycin, clarithromycin, dirithromycin, and roxithromycin.
5Trimethoprim-sulfamethoxazole testing predicts susceptibility and resistance to trimethoprim-sulfamethoxazole and sulfonamides.
6The Mueller-Hinton agar used for this test should be thymidine free to obtain accurate results.
7Trimethoprim-sulfamethoxazole is a combination of two drugs used in treatment (co-trimoxazole). The 1:20 ratio is that at which the greatest synergy in treatment has been demonstrated in serum. The disks are impregnated with 1.25 µg trimethoprim and 23.75 µg sulfamethoxazole to mimic the 1:20 ratio. The MIC breakpoints mimic the 1/20 ratio; i.e., ≤2 µg/ml trimethoprim/38 µg/ml sulfamethoxazole.
8Interpretations valid for other tetracyclines.
9The screening of pneumococci with reduced susceptibility to fluoroquinolones is realized by measuring sensitivity to norfloxacin. If the diameter around the disk of norfloxacin (5 ug) is less than 10 mm and/or if the MIC is > 16 µg/ml, there is a high risk of in vivo selection of mutants resistant to fluoroquinolones and clinical failure.
10Resistance to telithromycin must be verified by retesting in the absence of CO2.
Table 14. Kirby-Bauer disk diffusion zone diameter and MIC interpretive standards for S. pneumoniae as recommended by EUCAST
Data is from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) website, http://www.eucast.org, version 1.3, December, 2010.
Antimicrobial
Agent/ClassDisk
ContentZone Diameter,
Nearest Whole mmMIC Interpretive
Standard (µg/ml)S R S R PENICILLINS1 Oxacillin (screen)1,2 1 µg ≥ 20 < 20 - - Benzylpenicillin1,2,3 - - ≤ 0.064 > 2 CEPHEMS Cefotaxime4,5
Ceftriaxone4,5-
--
-≤ 0.5
≤ 0.5> 2
> 2MACROLIDES Erythromycin6 15 µg ≥ 22 < 19 ≤ 0.25 > 0.5 FOLATE PATHWAY INHIBITORS Trimethoprim-sulfamethoxazole (cotrimoxazole)7 1.25/23.75 µg ≥ 18 < 15 ≤ 1 >2 TETRACYCLINES Tetracycline8 30 µg ≥ 23 < 20 ≤ 1 > 2 FLUOROQUINOLONES9 Levofloxacin9,10 5 µg ≥ 19 < 19 ≤ 2 > 2 LINCOSAMIDES Clindamycin11 2 µg ≥ 19 < 19 ≤ 0.5 > 0.5 KETOLIDES Telithromycin10 15 µg ≥ 25 < 22 ≤ 0.25 > 0.5
Footnotes
1Most MIC values for penicillin, ampicillin, amoxicillin, and piperacillin (with or without a β-lactamase inhibitor) differ by no more than one dilution step and isolates fully susceptible to benzylpenicillin (MIC ≤0.064 µg/ml; susceptible by oxacillin disk screen, see note 2) can be reported susceptible to β-lactam agents that have been given breakpoints.
2Screen for β-lactam resistance with the oxacillin 1 µg disk. Isolates categorized as susceptible can be reported susceptible to benzylpenicillin, phenoxymethylpenicillin, and aminopenicillins (with or without β-lactamase inhibitor) irrespective of clinical indication. Isolates categorized as oxacillin resistant can be reported resistant to benzylpenicillin and phenoxymethylpenicillin in meningitis. For other β-lactams, determine the MIC of the agent considered for clinical use.
3In meningitis, only isolates with MIC ≤0.064 µg/ml (susceptible by oxacillin disk screen, see note 2) should be categorized susceptible to benzylpenicillin, otherwise report resistant. For indications other than meningitis and pneumonia, use breakpoints of 0.064 and 2 µg/ml.
4Strains with MIC values above the susceptible breakpoint are very rare or not yet reported. The identification and antimicrobial susceptibility tests on any such isolate must be repeated and if the result is confirmed the isolate sent to a reference laboratory. Until there is evidence regarding clinical response for confirmed isolates with MIC above the current resistant breakpoint, they should be reported resistant.
5Screen for β-lactam resistance with the oxacillin 1 µg disk. Isolates categorized as susceptible can be reported susceptible to cefepime, cefotaxime, cefpodoxime, ceftriaxone and cefuroxime, and cefuroxime axetil. Isolates categorized as oxacillin resistant should be tested with an MIC method with the agent considered for clinical use.
6Erythromycin can be used to determine susceptibility to azithromycin, clarithromycin, and roxithromycin.
7Trimethoprim-sulfamethoxazole in the ratio 1:19. Breakpoints are expressed as the trimethoprim concentration.
8Isolates susceptible to tetracycline are also susceptible to doxycycline and minocycline. Some isolates resistant to tetracycline may be susceptible to minocycline and/or doxycycline.
9The norfloxacin disk diffusion test can be used to screen for fluoroquinolone resistance. Isolates categorized as susceptible can be reported susceptible to levofloxacin and moxifloxacin and intermediate to ciprofloxacin and ofloxacin. Isolates categorized as resistant should be tested for susceptibility to individual agents.
10The breakpoints for levofloxacin relate to high dose therapy.
11Inducible clindamycin resistance can be detected only in the presence of a macrolide antibiotic. In disk diffusion tests, look for apparent antagonism of clindamycin by erythromycin (D-test).
standardization and QC should be conducted at each laboratory in accordance with the standardized guidelines presented in this manual.
Methods for preservation and storage of isolates and detailed methods for transport of isolates according to international regulations are presented in Chapter 14: Storage and Shipping of N. meningitidis, S. pneumoniae, and H. influenzae.
With increasing antimicrobial resistance testing being performed outside of international reference laboratories, antimicrobial gradient test strips serve as an assessment method that is both convenient and reliable. They require less technical expertise than MIC testing by dilution methods, but give comparable results. Store the antimicrobial gradient test strips as recommended by the manufacturer, usually at 4°C or in a freezer at -20°C.
Although this manual serves as a general guide for the use of antimicrobial gradient strips, always follow the manufacturer’s directions, as certain antibiotic-bacteria (“drug-bug”) combinations have special testing requirements.
This laboratory manual therefore suggests that Mueller-Hinton agar with 5% sheep blood should be used when performing antimicrobial susceptibility testing of S. pneumoniae with the antimicrobial test strips (except when testing for susceptibility to trimethoprim-sulfamethoxazole (cotrimoxazole), in which case horse blood should be used in place of sheep blood). Either 100-mm or 150-mm plates can be used, depending on the number of antimicrobial test strips used per sample (Figures 4 and 5). Two different antimicrobial test strips can be placed in opposite gradient directions on a 100-mm plate. This laboratory manual suggests that in order to avoid overlapping zones of inhibition of growth, not more than five antimicrobial test strips be used on a 150-mm plate. Depending on the bacteria/antimicrobial combinations, five strips on a 150 mm plate may lead to overlapping ellipses. If this occurs, redo the testing using four strips per plate.
Isolates to be tested should be subcultured onto a blood agar plate and incubated in a CO2-enhanced atmosphere (5% CO2 in a CO2-incubator or candle-extinction jar) at 35±2°C for 20-24 hours prior to testing. If the organism has been frozen, it should be subcultured twice when it is removed from the freezer before proceeding with susceptibility testing.
Warm the cation-adjusted Mueller-Hinton broth (CAMHB) to 35°C before using. Allow the gradient strips that will be used in the batch of testing to warm to room temperature (25°C). It is recommended to follow the directions on the package insert included withthe gradient strips.
- Using a sterile cotton-tip applicator, touch the surface of one to four morphologically identical, isolated colonies. Immerse the applicator into a tube containing sterile CAMHB. Rub the applicator against the wall of the tube slightly to release a small amount of growth into the liquid. Cap the tube and mix the cells using a vortex to form a suspension, being careful not to form froth or bubbles in the suspension when mixing the cells. Adjust the turbidity of the inoculum to that of a 0.5 McFarland turbidity standard (approximately 1 to 4 x 108 CFU/ml). Preparation of aMcFarland turbidity standard is described inthe Annex. If the turbidity of the inoculum is greater than the standard, dilute it with CAMHB to equal the turbidity of the standard. This suspension must be used within 15 minutes.
- Perform regular colony counts to verify that the density of the inoculum suspension is correct. For example, dilute the suspension 1:100 and subculture 10 µl onto the recommended media. An acceptable inoculum should give approximately 100-500 colonies. It is not necessary to perform colony counts on every isolate tested.
- Allow the plate to dry for up to 15 minutes. Be sure the plate is entirely dry before proceeding. While the plate is drying, remove the antimicrobial test strips from cold storage (4°C or -20°C, depending on the manufacturer’s recommendations), and allow the strips that will be used in the batch of testing to warm to room temperature (25°C). Return the strips that will not be used in this batch of testing to cold storage.
- Place the antimicrobial test strips onto the dried, inoculated agar plate with an applicator or sterile forceps (Figures 4 and 5). Make sure that the printed MIC values are facing upward (i.e., that the bottom surface of the strip containing the antimicrobial gradient is in contact with the agar). Alternatively, robotic gradient strip applicators are available from some manufacturers. Once applied, do not move the antimicrobial gradient strips as the drug diffuses into the agar immediately on contact.
- Incubate the plates in an inverted position in a CO2-enriched atmosphere (2-5% CO2) for 20–24 hours at 37°C. A candle-extinction jar may be used if a CO2 incubator is not available.
- Always follow the manufacturer’s instructions included with each package of strips, because incubation conditions may vary by organism-antimicrobial (or “drug-bug”) combination.
- After incubation, there will be an ellipse of bacterial growth formed on the plate around the strip and the test strip can be read. It is important to review QC results before reading and interpreting the antimicrobial test strips MIC.
Reading and interpreting the gradient strips
Read the MIC at the point where the zone of inhibition intersects the MIC scale on the strip. Use oblique light to carefully examine the end point. A magnifying glass may be used if needed. Read at the point of complete inhibition of all growth including hazes. A reading guide for the gradient strips, which shows organism related effects, drug-related effects, resistance mechanism-related effects, and technical and handling effects can be found at: http://www.abbiodisk.com/pdf/pi/75002206.pdf [1.68 MB, 2 pages].
Record the QC results first. If zones produced by the control strain are out of the expected ranges (see Tables 2, 3, and 5, depending on the strains and guidelines being used), the laboratorian should consider possible sources of error. Because antimicrobial susceptibility test results can be affected by many factors not necessarily associated with the actual susceptibility of the organism (e.g., inoculum size, growth phase, agar depth, storage, time, and others), QC practices must be followed carefully (see Section II above). If all antimicrobial agents are in the control range, read the test MICs. Note any unusual observations such as a zone of incomplete killing (trailing endpoints) or single colonies growing within the ellipse.
The gradation marks on the gradient strip correspond to the standard 2-fold dilution concentrations for the agar dilution method, but also include increments between those standard values. The standard values are used for interpretation and reporting of antimicrobial susceptibility test results. It is advised that both the actual reading of the value from the strip and the next-higher standard value (i.e., the value to be used for interpretation) be included in the laboratory records for testing of the strain. For example, if testing susceptibility of an isolate to penicillin, an MIC recorded from the gradations on the gradient strip might be 0.094 µg/ml; however, the reported MIC would be 0.125 µg/ml.
The manufacturer of the gradient strips recommends following the MIC breakpoints developed for agar and broth microdilution. Interpretive breakpoints for S. pneumoniae antimicrobial combinations from CLSI (Table 12), CA-SFM (Table 13), and EUCAST (Table 14) are shown.
- Using a sterile cotton-tip applicator, touch the surface of one to four morphologically identical, isolated colonies. Immerse the applicator into a tube containing sterile CAMHB. Rub the applicator against the wall of the tube slightly to release a small amount of growth into the liquid. Cap the tube and mix the cells using a vortex to form a suspension, being careful not to form froth or bubbles in the suspension when mixing the cells. Adjust the turbidity of the inoculum to that of a 0.5 McFarland turbidity standard (approximately 1 to 4 x 108 CFU/ml). Preparation of aMcFarland turbidity standard is described inthe Annex. If the turbidity of the inoculum is greater than the standard, dilute it with CAMHB to equal the turbidity of the standard. This suspension must be used within 15 minutes.
-
References
- Alcala, B., C. Salcedo, L. de la Fuente, L. Arreaza, M. J. Uria, R. Abad, R. Enriquez, J. A. Vazquez, M. Motge, and J. de Batlle. 2004. Neisseria meningitidis showing decreased susceptibility to ciprofloxacin: first report in Spain. Journal of Antimicrobial Chemotherapy 53:409.
- Barry, A. L., S. D. Brown, and P. C. Fuchs. 1996. In vitro activity of ceftizoxime, ceftazidime, cefuroxime, ceftriaxone, cefotaxime, and penicillin against Streptococcus pneumoniae as determined by three quantitative methods. European Journal of Clinical Microbiology and Infectious Diseases 15:344-346.
- Billal, D. S., M. Hotomi, and N. Yamanaka. 2007. Can the Etest correctly determine the MICs of beta-lactam and cephalosporin antibiotics for beta-lactamase-negative ampicillin-resistant Haemophilus influenzae? Antimicrobial Agents and Chemotherapy 51:3463-3464.
- Chu, Y. W., T. K. M. Cheung, V. Tung, F. Tiu, J. Lo, R. Lam, R. Lai, and K. K. Wong. 2007. A blood isolate of N. meningitidis showing reduced susceptibility to quinolones in Hong Kong. International Journal of Antimicrobial Agents 30:94-95.
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; 18th Informational Supplement. CLSI document M100-S19. Wayne, P. C. L. S. I. 2011. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. , CLSI Document M100-S21 ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- Corso, A., D. Faccone, M. Miranda, M. Rodriguez, M. Regueira, C. Carranza, C. Vencina, J. A. Vazquez, and M. Galas. 2005. Emergence of Neisseria meningitidis with decreased susceptibility to ciprofloxacin in Argentina. Journal of Antimicrobial chemotherapy 55:596-597.
- Enriquez, R., R. Abad, C. Salcedo, and J. A. Vazquez. 2009. Nalidixic acid disk for laboratory detection of ciprofloxacin resistance in Neisseria meningitidis. Antimicrobial Agents and Chemotherapy 53:796-797.
- Fiebelkorn, K. R., S. A. Crawford, and J. H. Jorgensen. 2005. Mutations in folP associated with elevated sulfonamide MICs for Neisseria meningitidis clinical isolates from five continents. Antimicrobial Agents and Chemotherapy 49:536-540.
- Jorgensen, J. H. 1994. Areas of recent emphasis of the National Committee for Clinical Laboratory Standards Subcommittee on Antimicrobial Susceptibility Testing. Advances in Experimental Medicine and Biology 349:61-65.
- Jorgensen, J. H., J. M. Swenson, F. C. Tenover, M. J. Ferraro, J. A. Hindler, and P. R. Murray. 1994. Development of interpretive criteria and quality control limits for broth microdilution and disk diffusion antimicrobial susceptibility testing of Streptococcus pneumoniae. Journal of Clinical Microbiology 32.
- Lucas, T. J. 1979. An evaluation of 12 methods for the demonstration of penicillinase. Journal of Clinical Pathology 32:1061-1065.
- Manchanda, V., and P. Bhalla. 2006. Emergence of non-ceftriaxone-susceptible Neisseria meningitidis in India. Journal of Clinical Microbiology 44:4290-4291.
- Mortensen, J. E., M. J. Gerrety, and L. D. Gray. 2006. 2006. Surveillance of antimicrobial resistance in Neisseria meningitidis from patients in the Cincinnati tristate region (Ohio, Kentucky, and Indiana). Journal of Clinical Microbiology 44:1592-1593.
- Murray, P. R., E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.). 2007. Manual of Clinical Microbiology, 9th ed. ASM Press, Washington, D.C.
- Park, C. H., J. S. Lopez, and C. B. Cook. 1978. Acidometric agar plate method for ampicillin susceptibility testing of Haemophilus influenzae. Antimicrobial Agents and Chemotherapy 13:318-320.
- Rainbow, J., E. Cebelinski, J. Bartkus, A. Glennen, D. Boxrud, and R. Lynfield. 2005. Rifampin-resistant meningococcal disease. Emerging Infectious Diseases 11:977-979.
- Sejvar, J. J., D. Johnson, T. Popovic, J. M. Miller, F. Downes, P. Somsel, R. Weyant, D. S. Stephens, B. A. Perkins, and N. E. Rosenstein. 2005. Assessing the risk of laboratory-acquired meningococcal disease. Journal of Clinical Microbiology 43:4811-4814.
- Skoczynska, A., J.-M. Alonso, and M.-K. Taha. 2008. Ciprofloxacin resistance in Neisseria meningitidis, France. Emerging Infectious Diseases 14:1322-1323.
- Stefanelli, P., C. Fazio, G. La Rosa, C. Marianelli, M. Muscillo, and P. Mastrantonio. 2001. Rifampicin-resistant meningococci causing invasive disease: detection of point mutations in the rpoB gene and molecular characterization of the strains. Journal of Antimicrobial Chemotherapy 47:219-222.
- Strahilevitz, J., A. Adler, G. Smollan, V. Temper, N. Keller, and C. Block. 2008. Serogroup A Neisseria meningitidis with reduced susceptibility to ciprofloxacin. Emerging Infectious Diseases 14:1667-1669.
- Taha, M.-K., M. L. Zarantonelli, C. Ruckly, D. Giorgini, and J.-M. Alonso. 2006. Rifampin-resistant N. meningitidis. Emerging Infectious Diseases 12:859-860.
- Vázquez, J. A., R. Enriquez, R. Abad, B. Alcalá, C. Salcedo, and L. Arreaza. 2007. Antibiotic resistant meningococci in Europe: Any need to act? FEMS Microbiology Reviews 31:64-70.
- Wu, H. M., B. H. Harcourt, C. P. Hatcher, S. C. Wei, R. T. Novak, X. Wang, B. A. Juni, A. Glennen, D. J. Boxrud, J. Rainbow, S. Schmink, R. D. Mair, M. J. Theodore, M. A. Sander, T. K. Miller, K. Kruger, A. C. Cohn, T. A. Clark, N. E. Messonnier, L. W. Mayer, and R. Lynfield. 2009. Emergence of ciprofloxacin-resistant Neisseria meningitidis in North America. New England Journal of Medicine 360:886-892.
- Page last reviewed: April 15, 2016
- Page last updated: March 15, 2012
- Content source:


 ShareCompartir
ShareCompartir