Chapter 9: Identification and Characterization of Haemophilus influenzae
Printer friendly version [19 pages]
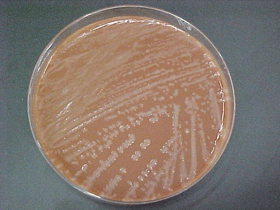
H. influenzae are small, pleomorphic, gram-negative bacilli or coccobacilli with random arrangements. H. influenzae is a fastidious organism which grows best at 35-37°C with ~5% CO2 (or in a candle-jar) and requires hemin (X factor) and nicotinamide-adenine-dinucleotide (NAD, also known as V factor) for growth. The standard medium used for growth of H. influenzae is a chocolate agar plate (CAP), which can be prepared with heat-lysed horse blood, a good source of both hemin and NAD, although sheep blood can also be used. Growth occurs on a CAP because NAD is released from the blood during the heating process of chocolate agar preparation (the heating process also inactivates growth inhibitors) and hemin is available from non-hemolyzed as well as hemolyzed blood cells. Alternatively, NAD can be included as a component of liquid H. influenzae growth media supplements, (available commercially or prepared in the laboratory), which are incorporated into the chocolate agar. H. influenzae appear as large, round, smooth, convex, colorless-to-grey, opaque colonies on a CAP (Figure 1). Encapsulated strains appear more mucoidal than non-encapsulated strains, which appear as smaller, compact grey colonies. No hemolysis or discoloration of the CAP is apparent. While H. influenzae produce a pungent indol smell, plates should not be opened in order to smell the cultures. H. influenzae cannot grow on an unsupplemented BAP. Prior to identification and characterization testing procedures, isolates should always be inspected for purity of growth and a single colony should be re-streaked, when necessary, to obtain a pure culture. For the following identification and characterization procedures, testing should be performed on 18-24 hour growth from a CAP at 35-37°C with ~5% CO2 (or in a candle-jar) (Figure 2).
The following tests are recommended to confirm the identity of cultures that morphologically appear to be H. influenzae (Figure 3). H. influenzae can be identified using Kovac’s oxidase test and determining the necessity of hemin and NAD as growth requirements. If the oxidase test is positive, hemin and NAD growth factor requirement testing should be performed. If the growth factor requirement test indicates that the isolate may be H. influenzae, serological tests to identify the serotype should be performed. This sequence of testing is an efficient way to save costly antisera and time. Additional methods for identification and characterization of H. influenzae using molecular tools are described in Chapter 10: PCR Methods and Chapter 12: Molecular Methods.
Biosafety Level 2 (BSL-2) practices are required for work involving isolates of H. influenzae, as this organism presents a potential hazard to laboratory personnel and the surrounding working environment. Please refer to Chapter 4: Biosafety in order to follow the guidelines that have been established for laboratorians working in BSL-2 facilities as many of the tests described in this chapter require opening plates with live cultures and are often performed outside of a biosafety cabinet (BSC).
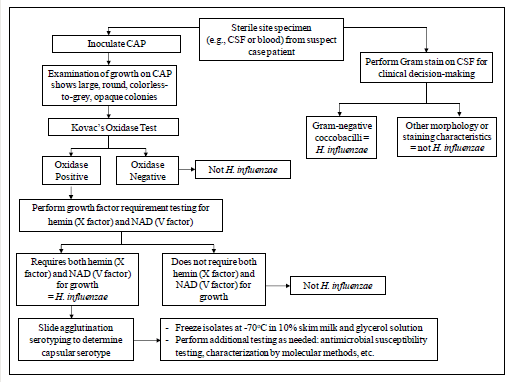
Figure 3. Flow chart for identification and characterization of a H. influenzae isolate...view larger
-
Kovac's oxidase test
Kovac’s oxidase test determines the presence of cytochrome oxidase. Kovac’s oxidase reagent, tetramethyl-p-phenylenediamine dihydrochloride, is turned into a purple compound by organisms containing cytochrome c as part of their respiratory chain. This test aids in the recognition of H. influenzae, but other members of the genus Haemophilus, as well as unrelated bacterial species, may also give a positive reaction. Positive and negative quality control (QC) strains should be tested along with the unknown isolates to ensure that the oxidase reagent is working properly.
-
Preparation of 1% oxidase reagent from oxidase powder
To prevent deterioration of stock oxidase powder, the powder should be stored in a tightly sealed desiccator and kept in a cool, dark area. Kovac’s oxidase reagent is intended only for in vitro diagnostic use. Avoid contact with the eyes and skin as it can cause irritation. In case of accidental contact, immediately flush eyes or skin with water for at least 15 minutes.
- Prepare a 1.0% Kovac’s oxidase reagent by dissolving 0.1 g of tetramethyl-p-phenylenediamine dihydrochloride into 10 ml of sterile distilled water.
- Mix well and then let stand for 15 minutes.
- The solution should be made fresh daily and the unused portion should be discarded.
- Alternatively, the reagent could be dispensed into 1 ml aliquots and stored frozen at -20°C. The aliquots should be removed from the freezer and thawed before use. Discard the unused portion each day the reagent is thawed.
-
Performing Kovac's oxidase test
Filter paper method
- Grow the isolate(s) to be tested for 18-24 hours on a CAP at 35-37°C with ~5% CO2 (or in a candle-jar).
- On a nonporous surface (i.e., Petri dish or glass plate), wet a strip of filter paper with a few drops of Kovac’s oxidase reagent.
- Let the filter paper strip air dry before use.
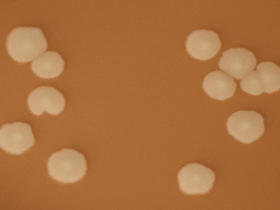
- Use a disposable plastic loop, a platinum inoculating loop, or a wooden applicator stick to pick a portion of a colony from overnight growth on the CAP and rub it onto the treated filter paper (Figure 4).
- Do not use a nichrome loop, as it may produce a false-positive reaction.
- Observe the filter paper for color change to purple.
- Perform steps 3 and 4 with a positive and negative QC strain to ensure that the oxidase reagent is working properly.
Plate method
- Grow the isolate(s) to be tested for 18-24 hours on a CAP at 35-37°C with ~5% CO2 (or in a candle-jar).
- Dispense a few drops of Kovac’s oxidase reagent directly on top of a few suspicious colonies growing on the 18-24 hour CAP.
- Do not flood the entire plate as the bacteria exposed to the reagent are usually not viable for subculture.
- Tilt the plate and observe colonies for a color change to purple.
- Perform steps 1 and 2 with a positive and negative QC strain to ensure that the oxidase reagent is working properly.
- Reading the oxidase test results
- Positive reactions will develop within 10 seconds in the form of a purple color where the bacteria were applied to the treated filter paper. Delayed reactions are unlikely with H. influenzae.
- Negative reactions will not produce a color change on the treated filter paper.
-
-
Identification of hemin and NAD as growth requirements
H. influenzae is a fastidious organism and can be identified on the basis of growth requirements for hemin and NAD. H. influenzae can be differentiated from most other species of Haemophilus by its specific requirement for both hemin and NAD for growth (Table 1). H. haemolyticus is the only other species requiring both hemin and NAD for growth; however, this species differs from H. influenzae by producing beta-hemolysis (clear) on horse or rabbit blood. For patients with bacterial meningitis, H. influenzae must be considered as the presumptive causative agent as opposed to H. haemolyticus when both hemin and NAD factors are required for growth. To differentiate between the two species, hemolysis must be checked on horse or rabbit blood agar (see Section II.B., Haemophilus ID Quad plate section below). H. haemolyticus usually causes hemolysis on these media, while H. influenzae does not. It has recently been reported that H. haemolyticus tend to rapidly lose their hemolytic property when passed in vitro (2). This has made the definitive identification of H. influenzae and H. haemolyticus using only biochemical tests very difficult and other methods, such as molecular testing, may be employed for differentiating between the two species.
Table 1. Identification of Haemophilus spp. by their growth requirements for hemin (X factor) and NAD (V factor) and β-hemolysis on horse blood agar
Organism Requirement for hemin (X factor) Requirement for NAD (V factor) β-hemolysis on horse blood agar H. influenzae + + - H. parainfluenzae1 - + - H. haemolyticus + + + H. parahaemolyticus - + + H. aphrophilus2 + - - H. paraphrophilus1,2 - + -
Footnotes
1H. parainfluenzae is ornithine decarboxylase positive, whereas H. paraphrophilus is negative.
2Although their requirements for hemin and NAD factor differ from each other, H. aphrophilus and H. paraphrophilus have recently been reclassified as a single species with a new genus: Aggregatibacter aphrophilus (3).
-
Performing hemin and NAD growth factor requirement test using paper disks and/or strips
Growth factor requirements can be identified with paper disks and/or strips using the principles of agar diffusion.
Growth factor requirement procedure using paper disks and/or strips
- Grow the isolate(s) to be tested for 18-24 hours on a CAP at 35-37°C with ~5% CO2 (or in a candle-jar).
- Prepare a moderately heavy suspension of cells (comparable to a 1.0 McFarland standard) from overnight growth on the CAP in a suitable broth (trypticase soy, heart infusion, or peptone water) and mix well using a vortex.
- Do not transfer any of the chocolate agar media from the plate to the cell suspension as even the smallest amount of agar will affect the test and may lead to misidentification of the bacteria.
- Inoculate one half of a heart infusion or trypticase soy agar plate with 10 µl of the cell suspension using a sterile loop or swab and allow the suspension to dry.
- Two different isolates can be tested on the same plate, but care must be taken to ensure that the cultures do not overlap.
- Place paper disks or strips containing hemin, NAD, and hemin/NAD on the inoculated plate after the inoculum has dried.
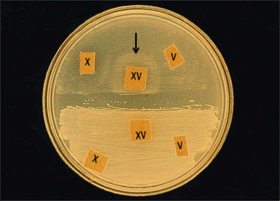
- When two bacterial strains are tested on the same plate (Figure 5), the disks must be placed in the exact manner shown, keeping the individual hemin and NAD disks separated by the one containing both factors and leaving as much space between the disks as possible.
- Perform steps 1-4 using a H. influenzae and a different Haemophilus spp. QC strain to ensure that the hemin and NAD disks or strips are working properly.
- Carefully invert the plate and incubate for 18-24 hours at 35-37°C with ~5% CO2 (or in a candle-jar).
- Observe growth around the paper disks or strips.
Reading the hemin and NAD paper disk and/or strip results
- H. influenzae will only grow around the paper disk containing both hemin and NAD, as shown in Figure 5 on the upper half of the plate (see black arrow).
- H. haemolyticus will also only grow around the paper disk containing both hemin and NAD. To differentiate between the two species, hemolysis must be checked on horse or rabbit blood agar by inoculating the cell suspension mentioned above on heart infusion agar with 5% rabbit blood (or agar infusion base containing horse blood). Alternatively, a Haemophilus ID Quad plate can be used (see Section II.B. below).
- Other Haemophilus spp. will grow around the disk containing both hemin and NAD and either the individual hemin or the NAD disk.
- Alternatively, the porphyrin test (2) can be used. This determines the hemin requirement of the isolate while avoiding the problem of hemin carryover from primary culture media and hemin contamination of test media (3).
-
Performing hemin and NAD growth factor requirement test using Haemophilus ID Quad plates
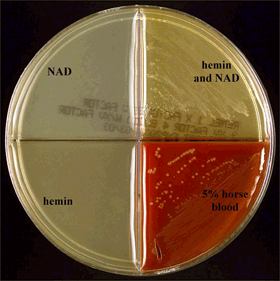
Haemophilus ID Quad plates are another method for determining growth requirements of Haemophilus isolates (Figure 6). While they are more expensive than the paper disks or strips, they can test for β-hemolysis (clear) on horse blood and assist in differentiating H. haemolyticus from H. influenzae. The Quad plate is divided into four compartments. One quadrant includes medium containing hemin only (Figure 6, lower left). The second quadrant includes medium containing NAD only (Figure 6, upper left). The third quadrant contains medium that includes both hemin and NAD (Figure 6, upper right). The fourth quadrant contains heart infusion agar or blood agar base with 5% horse blood (Figure 6, lower right) for detecting hemolysis and differentiating H. haemolyticus from H. influenzae.
Growth factor requirement procedure using Haemophilus ID Quad plates
- Grow the isolate(s) to be tested for 18-24 hours on a CAP at 35-37°C with ~5% CO2 (or in a candle-jar).
- Prepare a suspension of cells (comparable to a 0.5 McFarland standard) from overnight growth of suspected Haemophilus on a CAP in trypticase soy broth or distilled water and mix well using a vortex.
- Use a sterile, inoculating loop to streak one loopful of the cell suspension onto one quadrant of the Quad plate. Streak the entire quadrant, starting at the periphery of the plate and streaking toward the center of the plate.
- Use a different inoculating loop to streak each of the other quadrants with the cell suspension.
- Stab into the blood agar for detection of β-hemolysis (clear).
- Incubate for 18-24 hours at 35-37°C with ~5% CO2 (or in a candle-jar).
- After incubation, examine the individual quadrants for growth and the quadrant with horse blood for hemolysis where the plate was stabbed with the loop (Figure 6).
Reading the Haemophilus ID Quad plate results
- H. influenzae will only grow on the quadrant containing both hemin and NAD and the quadrant containing horse blood. It will not hemolyze the horse blood cells.
- H. haemolyticus may lose their hemolytic property when passed in vitro. This has made the definitive identification of H. influenzae and H. haemolyticus using only biochemical tests very difficult and other methods, such as molecular testing, may be employed for differentiating between the two species.
- An organism growing on either the hemin or the NAD quadrant is likely another Haemophilus species
- If growth occurs on every quadrant, the isolate is probably not a Haemophilus spp.
- H. influenzae may occasionally show slight growth in the quadrant containing NAD only.
Quality control of Quad plates
QC should be performed on each new lot of Quad plates before they are used for unknown isolates to ensure that they will support the proper growth of Haemophilus spp. Three plates from each new lot received should be tested using a well-characterized reference strain of H. influenzae, H. haemolyticus, and H. parahaemolyticus. One uninoculated plate from each new lot should also be tested in order to check for contamination of mold or other organisms in the laboratory and/or incubator. QC should be repeated on plates from a lot if they have been exposed to temperatures above 4°C or if there is reason to suspect that the plates have been contaminated since the initial QC was performed.
Procedure for quality control of Quad plates
- Examine the Quad plates for evidence of microbial contamination, discoloration, drying, deterioration, or other physical defects that may interfere with use. Note the firmness of the agar during the inoculation procedure.
- Grow the reference strains to be tested for 18-24 hours on a CAP at 35-37°C with ~5% CO2 (or in a candle-jar).
- Inoculate the Quad plates using cell suspensions from the reference strains as described in the Quad plate procedure above.
- Incubate the plates for 18-24 hours at 35-37°C with ~5% CO2 (or in a candle-jar).
- As a negative control for contamination, incubate an uninoculated plate from each new lot for 18-24 hours at 35-37°C with ~5% CO2 (or in a candle-jar).
- Examine the inoculated and uninoculated plates after 18-24 hours for proper growth of Haemophilus spp.
Reading the quality control test results
- H. influenzae will only grow on the quadrant containing both hemin and NAD and the quadrant containing horse blood. It will not hemolyze the horse blood cells.
- H. haemolyticus will only grow on the quadrant containing both hemin and NAD and the quadrant containing horse blood. It will hemolyze the horse blood cells.
- H. parahaemolyticus will grow on all quadrants except for the one containing only hemin. It will hemolyze the horse blood cells.
- H. parahaemolyticus will grow on all quadrants except for the one containing only hemin. It will hemolyze the horse blood cells.
- Failing result: no growth or poor growth of the reference strain on appropriate media and/or growth of organisms on the uninoculated media.
-
-
Identification of H. influenzae serotype
Haemophilus influenzae can be encapsulated with one of six types of antigenically distinct capsules which can be serotyped using antisera to each capsule (serotypes a-f). H. influenzae may also be non-encapsulated and such strains that cannot be serotyped are called H. influenzae nontypeable (NT). Individual serotype-specific antisera for these major serotypes are available commercially. A polyvalent antiserum that recognizes all 6 serotypes is also available commercially.
It is not always practical to test for all serotypes for which antisera are available in a laboratory. Testing algorithms may be set up in laboratories with previous knowledge of whether or not a H. influenzae serotype b (Hib) vaccination program has been implemented within that particular geographic region. Modifications may be made to the testing algorithm for any laboratory based on information regarding Hib vaccination status of the region.
It is essential that reference laboratories have the capacity to isolate, identify, and characterize the serotype of isolates of H. influenzae. This valuable data provides laboratories and public health authorities with the tools to identify outbreaks controllable by vaccination campaigns and recognize serotypes causing sporadic disease.
SAST test algorithm for areas without an established Hib vaccination program
If a Hib vaccination program has not been implemented in the country or region from which the isolate originated, it is likely that the H. influenzae isolate is serotype b and the isolate should first be tested for reactivity to serotype b antisera and a negative saline control. If the isolate reacts positively with the serotype b antiserum with no agglutination in saline, the isolate is identified as Hib. However, if the isolate is non-reactive with the serotype b antiserum, and if polyvalent antiserum is available, it should be tested with the polyvalent antiserum. If positive, the isolate should then be tested with the remaining monovalent antisera (a, c, d, e, and f) to determine the serotype. If negative for all monovalent antisera and positive for hemin and NAD growth requirements, then the isolate is considered NT.
SAST test algorithm for areas with an established Hib vaccination program
If the isolate is from a country or region with an established Hib vaccination program that has high Hib vaccine coverage, the isolate is likely to be NT or a serotype other than b. In this case, the isolate should first be tested with the polyvalent antiserum, if available, and a negative saline control. If positive for the polyvalent antiserum with no agglutination in the saline, the isolate should then be tested with the remaining monovalent antisera (a, b, c, d, e, and f) to determine the serotype. If the isolate is negative for the polyvalent antiserum and/or the monovalent antisera, and requires hemin and NAD for growth, then the isolate is considered NT.
-
Slide agglutination serotyping (SAST) test for serotyping H. influenzae isolates
Formalin-killed suspensions of H. influenzae should be used for SAST testing rather than saline suspensions of living organisms to maintain a safe working environment. A solution of 5% formalinized physiological saline is sufficient to kill the bacteria. However, formalin is a carcinogen and must be stored and handled with great care. Alternatively, work should be performed under a biosafety hood if formalin is not used. Antisera should be stored in the refrigerator at 4°C and warmed to room temperature (25°C) before use. It must be put back in the refrigerator as soon as testing is finished to prevent the loss of binding activity of the antibody.
- Performing the SAST test
- Grow the isolate(s) to be tested for 18-24 hours on a CAP at 35-37°C with ~5% CO2 (or in a candle-jar).
- Clean a glass slide with alcohol (optional if slides are pre-cleaned).
- Divide the slide into equal sections (e.g., twelve 11 X 22 mm sections on a standard 50 X 75 mm slide) with a liquid impermeable pen or a wax pencil.
- Each isolate will require as many sections on the slide as antisera that will be tested (polyvalent and/or individual serotype-specific) as well as a saline negative control.
- In the lower portion of each of the sections of the glass slide described in step (2), add 10 µl of the 5% formalinized saline with a micropipettor.
- The instructions specify using a micropipettor with sterilized filtered tips to measure the 10 µl of the 5% formalinized saline to suspend the bacteria. The micropipettor will transfer precise and equal measurements for a proper SAST reaction.
- If a micropipettor and tips are not available, sterile, disposable 10 µl inoculation loops can be used to transfer 10 µl of the 5% formalinized saline, but often do not deliver accurate amounts (between 5-10 µl).
- Use a sterile, disposable 10 µl inoculating loop to collect a few colonies from the surface of the overnight culture incubated on the CAP.
- Suspend the bacteria in the 5% formalinized saline solution in the lower portion of each of the sections of the slide. The suspension should be moderately opaque (see saline control in Figure 7). Do not allow the cell suspension to dry before adding the antisera.
- If the bacteria are difficult to suspend directly on the slide, make a moderately milky suspension (comparable to McFarland 6.0 standard) of the test culture in a small vial with 250 µl of 5% formalinized saline and briefly vortex the suspension to mix and break up any pellets. Add 10 µl of this suspension to the lower portion of the slide.
- In the upper portion of each of the sections of the glass slide described in step (2), add 10 µl of the polyvalent and/or serotype-specific antisera to be tested as well as unformalinized saline or phosphate buffered saline (PBS) for a negative control with a micropipettor.
- DO NOT use the dropper provided with the antisera because it usually delivers larger amounts than is necessary and can easily be contaminated.
- If a micropipettor and tips are not available, sterile, disposable 10 µl inoculation loops can be used to transfer 10 µl of the antisera, but often do not deliver accurate amounts (between 5-10 µl).
- Dispose of the tip or loop used to transfer the antisera to the slide in a waste container after each use to avoid contamination of the antisera. If the source of antisera is contaminated, a new vial must be used.
- Gently tilt the slide to mix the cell suspensions with the antisera in each section. Continue to gently rock the slide for 1 to 2 minutes to allow the lower and upper portions to completely blend. Do not use a circular motion while rocking, as it can cause the sections with different serogroup-specific antisera to run together and contaminate each other.
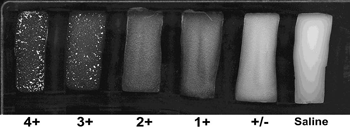
- After 2 minutes, examine the SAST reactions under a bright light and over a black background. Use the rating system in Figure 7 to determine the intensity of the agglutination reaction in each section of the slide. Disregard any agglutination that occurs after the 2 minute time period.
- Record the SAST results in the laboratory log book.
-
Reading the SAST results
Rating the intensity of the agglutination reaction
Agglutination occurs when the antisera bind to the bacterial cells causing the cells to agglutinate or clump together, thus making the cell suspension appear clearer. The intensity of the agglutination reaction may vary according to the density of the cell suspension or the antisera used. A description on the intensity ratings shown in Figure 7 are listed below.
4+ All of the cells agglutinate and the cell suspension appears clear
3+ 75% of the cells agglutinate and the cell suspension remains slightly cloudy
2+ 50% of the cells agglutinate and the cell suspension remains slightly cloudy
1+ 25% of the cells agglutinate and the cell suspension remains slightly cloudy
+/- Less than 25% of the cells agglutinate and a fine granular matter occurs
0 No visible agglutination; the suspension remains cloudy and smoothDetermining the serotype
- A positive result is designated by a 3+ or 4+ (strong agglutination) within 1-2 minutes.
- A negative result is designated by a 0 (saline), +/-, 1+ or 2+ (weak agglutination).
- The serotype is determined when a positive result occurs with the polyvalent antiserum and/or only one of the serotype-specific antisera and not with the saline.
- If a serotype is not determined, the isolate is considered NT. The following result combinations are all reported as NT:
- Agglutination in the saline, regardless of strong reactions with the polyvalent or other serotype-specific antisera, characterizes the culture as autoagglutinating.
- Agglutination with the polyvalent and/or more than one serotype-specific antisera in the absence of agglutination in saline characterizes the culture as polyagglutinating or cross-reactive.
- No agglutination with the polyvalent or any of the serotype-specific antisera or the saline characterizes the strain as non-reactive.
- Although rare, an isolate positive for the polyvalent antiserum, but negative for the serotype-specific antisera is considered NT.
-
Troubleshooting the SAST procedure
H. influenzae isolates are subject to variability (encapsulated vs. unencapsulated, small vs. large colonies, slow growers vs. fast growers, and heavy agglutinators vs. light agglutinators) and may be unclear or difficult to interpret. Some troubleshooting procedures are listed below:
- Repeat the test directly on the slide using growth from another section of the same plate.
- Make a cell suspension in a small tube and vortex if the result from SAST directly on the slide is unclear and repeat the test.
- Add 20 µl of antisera directly to slide and then add a loopful of organism without diluting the specimen with 5% formalinized saline.
- Subculture and retest fresh growth the following day.
- If the original plate contains different size colonies, make a subculture for each type of colony and test both cultures the next day. The larger colonies usually indicate better capsule production and therefore better reactivity. However, the smaller colonies will occasionally give a better result.
- If discrepancies are not immediately resolved, any subsequent SAST repeats should be used in conjunction with control strains.
-
Quality control of antisera for SAST testing
A set of reference strains for H. influenzae serotypes a, b, c, d, e, and f (one per serotype) and a nontypeable H. influenzae strain should be used to QC the antisera before testing any unknown isolates. QC of the antisera should be:
- Performed for each new lot of antisera received in the laboratory.
- Performed biannually after initial QC testing.
- Repeated if a vial has been exposed to temperatures above 4°C or if there is reason to suspect that the vial has been contaminated since the initial QC was performed.
Follow the SAST testing procedure to QC each lot of antisera using all reference strains available in the laboratory. Record the results provided on the example QC sheet in Figure 8.
Reading the quality control test results
Passing test:
- The antiserum must give 3+ or 4+ agglutination with homologous antigens within 1-2 minutes.
- The antiserum must not react with heterologous H. influenzae serotypes, with the NT reference strain, or in saline.
Failing test:
- The antiserum agglutinates with one or more reference strains and/or with the NT reference strain and/or in saline.
Figure 8. Example QC sheet for testing antisera against all H. influenzae serotypesA
(strain number)B
(strain number)C
(strain number)D
(strain number)E
(strain number)F
(strain number)NT
(strain number)A
Lot#++++ - - - - - - B
Lot#- ++++ - - - - - C
Lot#- - ++++ - - - - D Lot# - - - ++++ - - - E
Lot#- - - - ++++ - - F
Lot#- - - - - ++++ - Poly
Lot#++++ ++++ ++++ ++++ ++++ ++++ - Saline - - - - - - -
-
-
Commercial identification kits
Several commercial identification systems that use biochemical or enzymatic substrates are available for identification of Haemophilus spp. These systems may occasionally require supplemental tests, and additional characteristics, such as microscopic and colony morphology, must be considered. Generally, each system is self-contained, but the addition of one or more reagents to complete certain reactions may be necessary. The manufacturer’s instructions should be followed precisely when using these kits. Some of the commercial identification kits also include biotyping of H. influenzae. For detailed instructions and use of appropriate control strains, consult the Clinical Microbiology Procedures Handbook (1).
References
- Clinical Microbiology Procedures Handbook, 3rd edition. 2010. ASM. Washington, D.C. 2,540 pages.
- Murray, P. R., E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.). Manual of Clinical Microbiology, 9th ed, vol. 2007. ASM Press, Washington, D. C.
- Norskov-Lauritsen, N., and M. Kilian. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include NAD factor-dependent and NAD factor-independent isolates. 2006. International Journal of Systematic and Evolutionary Microbiology. 56:2135-2146.
- Page last reviewed: April 15, 2016
- Page last updated: March 15, 2012
- Content source:


 ShareCompartir
ShareCompartir