Diagnosis and Treatment
How Can Salmonella Infections Be Diagnosed?
Diagnosing salmonellosis requires testing a clinical specimen (such as stool or blood) from an infected person to distinguish it from other illnesses that can cause diarrhea, fever, and abdominal cramps. Once Salmonella is identified in the specimen, additional testing can be done to further characterize the Salmonella.
Steps in Laboratory Testing and Reporting Salmonella
- Laboratory scientists identify Salmonella infection by culturing a patient’s sample. If Salmonella bacteria grow, then the diagnosis is confirmed, or in laboratory-terms, “culture confirmed.”
- Clinical diagnostic laboratories report the test results to the treating clinician and submit Salmonella isolates to state and territorial public health laboratories for serotyping and DNA fingerprinting.
- The public health laboratories report the results to CDC’s Laboratory-based Enteric Disease Surveillance and to PulseNet
- The public health laboratories forward atypical serotypes to CDC’s National Salmonella Reference Laboratory for more characterization or confirmation.
Serotyping Salmonella
Serotype: group within a single species of microorganisms, such as bacteria or viruses, which share distinctive surface chemical structures
Culture: Growing bacteria, viruses, and other living matter in a specific environment, such as a petri dish coated with nutrients to encourage growth
Salmonella are divided into serotypes according to structures on the bacteria’s surface. Serotyping is used in outbreak investigations to link cases of illness with similar bacteria and track them to the source (example: a contaminated food or an infected animal). Some serotypes are only found in one kind of animal or in a single place. Others are found in many different animals and all over the world. Some serotypes can cause especially severe illnesses when they infect people; most typically cause milder illnesses.
Learn more about the importance of serotyping and CDC’s Salmonella Atlas, the first-of-its-kind report that charts over 40 years of laboratory-confirmed surveillance data on 32 Salmonella serotypes.
Serotyping has played an important role in the understanding the epidemiologic and molecular characterization of Salmonella for decades. Today, modern genetic subtyping methods provide scientists with additional information to understand common serotypes and identify, investigate, and trace outbreaks.
PulseNet and Pulsed-Field Gel Electrophoresis (PFGE)
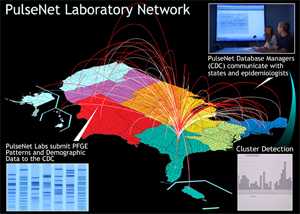
In addition to serotyping, state public health laboratories routinely subtype Salmonella isolates using pulsed-field gel electrophoresis (PFGE) to create a “DNA fingerprint.”
The laboratories submit these fingerprint patterns to a dynamic database maintained by PulseNet, a national network of public health and food regulatory agency laboratories coordinated by CDC. The network includes state health departments, local health departments, and federal agencies (CDC, United States Department of Agriculture’s Food Safety and Inspection Service (USDA/FSIS), and the US Food and Drug Administration (FDA)).
PulseNet data are available to participating health departments for comparing PFGE patterns. Finding a group of infections with the same pattern could indicate an outbreak. Finding the same pattern in a food could help link illness to a specific food source. View PulseNet’s PFGE protocol for Salmonella isolates [PDF - 13 pages].
Sequence-based subtyping with MLVA
In addition to PFGE, Multiple Locus Variable-number Tandem Repeat Analysis, or MLVA, is another technique used by PulseNet to further characterize Salmonella isolates. See the table below for a complete list of PulseNet’s laboratory and analysis procedures for MLVA.
Next Generation of Laboratory Testing
 New methods for subtyping. PulseNet is working with other Federal and State Agencies to evaluate the use of whole genome sequencing (WGS) to replace PFGE as its standard method for subtyping Salmonella and other bacteria that cause diarrheal disease. It is hoped that WGS will make the PulseNet process faster, more accurate, and more efficient.
New methods for subtyping. PulseNet is working with other Federal and State Agencies to evaluate the use of whole genome sequencing (WGS) to replace PFGE as its standard method for subtyping Salmonella and other bacteria that cause diarrheal disease. It is hoped that WGS will make the PulseNet process faster, more accurate, and more efficient.
New methods for diagnosis. CDC is also working to adapt its processes to important changes in how Salmonella is diagnosed in patients. Until recently, culture (laboratory procedures to grow living Salmonella from patient specimens) was the diagnostic standard and the cornerstone of public health surveillance.
The Food and Drug Administration (FDA) has recently approved new culture independent diagnostic testing (CIDT) methods for diagnosing Salmonella infections, but they are not widely used yet. These new methods do not depend on culturing the specimen, so they do not yield an isolate for serotyping or PFGE. When adopted, these new tests could impact surveillance in a number of ways. PulseNet hopes to develop sequence-based tests that are themselves culture-independent using new systems developed for WGS. View a comparison of culture-dependent and culture-independent methods.
Additional Resources
- Learn more about the Kauffmann-White scheme used to identify serotypes.
- Learn more about national Salmonella surveillance data by serotype going back to 1963.
- Page last reviewed: March 9, 2015
- Page last updated: March 9, 2015
- Content source:


 ShareCompartir
ShareCompartir