Notes from the Field: Investigation of Elizabethkingia anophelis Cluster — Illinois, 2014–2016
Weekly / December 9, 2016 / 65(48);1380–1381
Livia Navon1,2; Whitney J. Clegg1; Jodi Morgan1; Connie Austin1; John R. McQuiston3; David D. Blaney3; Maroya Spalding Walters4; Heather Moulton-Meissner4; Ainsley Nicholson3 (View author affiliations)
View suggested citationElizabethkingia spp., formerly known as Flavobacterium and Chryseobacterium, are multidrug-resistant, Gram negative bacilli found in the environment that can cause health care–associated outbreaks (1). Elizabethkingia meningoseptica was first identified by Elizabeth King in 1959 as a cause of meningitis outbreaks among hospitalized newborns (2). Elizabethkingia anophelis (EKA) was first identified in 2011 from the midgut of a mosquito (3); a recent series of cases from Hong Kong indicate that EKA health care–associated infections cause significant morbidity and have a high case-fatality rate (23.5%) (4).
In February 2016, the Wisconsin Department of Health Services notified the Illinois Department of Public Health (IDPH) and other neighboring health departments of an ongoing outbreak of EKA among Wisconsin residents. To determine if Illinois had related cases, IDPH sent memos on February 10 and March 29, 2016 to Illinois health care providers, infection preventionists and laboratories, requesting all available isolates of Elizabethkingia spp. dating back 2 years, to January 1, 2014. Twelve isolates from 11 patients were sent to CDC for testing; specimen collection dates ranged from June 23, 2014 to March 31, 2016.
On April 14, 2016, CDC informed IDPH that all submitted isolates were identified as EKA and that a genetic cluster (11 isolates from 10 patients) distinct from the Wisconsin outbreak strain had been identified, based on pulsed-field gel electrophoresis (PFGE) and whole genome sequencing (WGS). The eleven isolates were an average of 39.6 single nucleotide polymorphisms (SNPs) apart by WGS, with a range of 9–60 SNPs in the core of the genomic sequence shared across the isolates (80% of the genome). This SNP range corresponded to PFGE patterns with zero (indistinguishable) to three (closely related) band pattern differences. By comparison, some historic EKA isolates tested by CDC have differed by approximately 1,000 SNPs, with the more distantly related EKA strains differing by tens of thousands of SNPs. Phylogenetic analysis followed by bootstrapping statistical analysis provided strong support that these Illinois isolates clustered together and were genetically distinct from other EKA isolates submitted to CDC.
During April–June 2016, IDPH conducted an investigation to identify risk factors and a potential source of infection among the 10 EKA cases. Cases were defined as the culture of EKA from sterile sites or the respiratory tract of Illinois patients from January 1, 2014 onward, and at least one specimen that was <60 SNPs distance by WGS to the cluster pattern identified by CDC. Eight patients had positive blood cultures and two had positive respiratory specimens.
Medical records of the 10 patients for the 30 days before collection of the EKA-positive specimen were reviewed. The median age at patient diagnosis was 68 years (range = 35–83 years), and seven of the patients were male. Patients resided in three nonneighboring counties in Northern Illinois. Comorbidities were common: nine patients had chronic obstructive pulmonary disease, eight patients had diabetes and seven patients had unhealed wounds. Eight patients were intubated and mechanically ventilated at the time of the first positive culture and seven patients had a percutaneous endoscopic gastrostomy tube in place. The case fatality rate was high: seven of the 10 patients died before June 2, including six who died within 30 days of positive EKA specimen collection.
In the 30 days preceding the first positive culture, all 10 patients resided in a health care facility and nine had received care at two or more facilities (Figure). Patients received inpatient care in a total of 19 facilities, including eight hospitals, seven nursing homes, and two long-term acute care hospitals. Facility overlap was limited; two facilities provided care to two patients each.
Because isolated Elizabethkingia spp. infections are not reportable to IDPH, baseline incidence data were not available. To determine whether the 10 identified cases represented an increase in EKA infections, the 19 facilities with patients in the cluster were asked to report all patients with Elizabethkingia spp. infections from January 1, 2012 to May 16, 2016. Fifteen facilities responded and reported a total of 77 patients, with an average of 17.1 infections per year (range = 13–19). The average number of infections per facility was 5.1 (range = 1–16).
Three outbreaks (2008, 2009, and 2012–2013) of Elizabethkingia meningoseptica have been reported previously in Illinois healthcare facilities. Environmental isolates collected and stored from the 2012–2013 outbreak were sent to CDC for testing to better understand the genetic diversity of Elizabethkingia spp. in Illinois. WGS indicated that the 2012–2013 environmental isolates were actually EKA and that their genomes clustered with the 2014–2016 case isolates by both PFGE and WGS.
The evidence does not support a finding that the recently identified cluster represents an acute, point source outbreak, given the lack of common facility exposure among patients, and that the number of infections in 2014–2016 reported by facilities did not appear to be higher than in previous years and the isolates from the 2014–2016 cluster matched environmental isolates from the 2012–2013 outbreak. Instead, this more likely represents ongoing sporadic infection among critically ill patients. Of note, after the investigation was completed, additional genetic analysis conducted by CDC indicated that one of the isolates initially identified as part of the cluster had a distinct PFGE pattern and by WGS, differed from the cluster genomes by 160–170 SNPs.
Molecular typing methods can identify clusters that might not be recognized by epidemiologic factors alone, and advanced techniques, such as WGS, can provide an additional level of discrimination compared with more established approaches, such as PFGE. However, molecular typing results must be interpreted cautiously, particularly for rare organisms for which there is limited information about mutation rates and genetic diversity. The findings of this cluster investigation emphasize that epidemiologic and clinical data remain critical to defining outbreaks and informing investigations.
Acknowledgments
Division of Laboratories, Illinois Department of Public Health; Wisconsin Department of Public Health; Wisconsin State Laboratory of Hygiene; Chicago Department of Public Health; infection preventionists at the facilities in which cases received care.
Corresponding author: Livia Navon, len8@cdc.gov, 312-814-3020.
1Illinois Department of Public Health; 2Division of State and Local Readiness, CDC; 3Division of High-Consequence Pathogens and Pathology, CDC; 4Division of Healthcare Quality Promotion, CDC.
References
- Jean SS, Lee WS, Chen FL, Ou TY, Hsueh PR. Elizabethkingia meningoseptica: an important emerging pathogen causing healthcare-associated infections. J Hosp Infect 2014;86:244–9. CrossRef PubMed
- King EO. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am J Clin Pathol 1959;31:241–7. CrossRef PubMed
- Kämpfer P, Matthews H, Glaeser SP, Martin K, Lodders N, Faye I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int J Syst Evol Microbiol 2011;61:2670–5. CrossRef PubMed
- Lau SK, Chow WN, Foo CH, et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep 2016;6:26045. CrossRef PubMed
 FIGURE. Patient* and specimen characteristics and health care facility exposures and outcomes 30 days before and after first positive specimen collection — Elizabethkingia anophelis (EKA) cluster, Illinois, 2014–2016
FIGURE. Patient* and specimen characteristics and health care facility exposures and outcomes 30 days before and after first positive specimen collection — Elizabethkingia anophelis (EKA) cluster, Illinois, 2014–2016
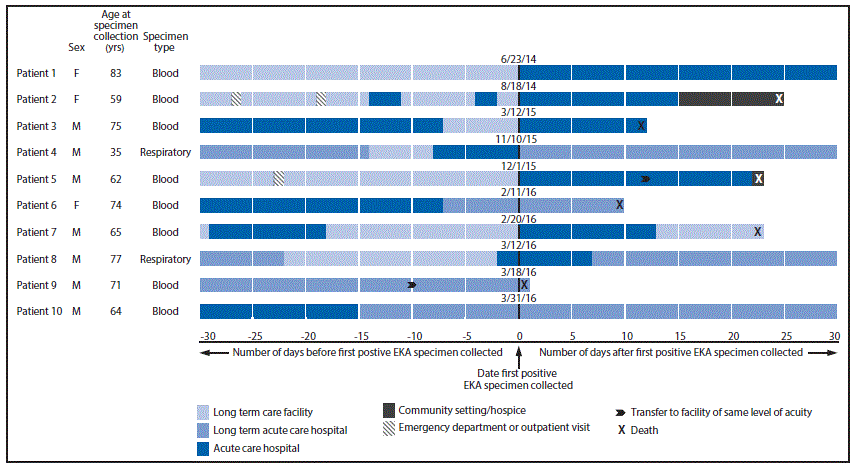
* After the investigation was completed, the isolate from patient 5 was determined to be 160–170 single nucleotide polymorphisms distance from the isolates in the genetic cluster.
Suggested citation for this article: Navon L, Clegg WJ, Morgan J, et al. Notes from the Field: Investigation of Elizabethkingia anophelis Cluster — Illinois, 2014–2016. MMWR Morb Mortal Wkly Rep 2016;65:1380–1381. DOI: http://dx.doi.org/10.15585/mmwr.mm6548a6.
Use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of
Health and Human Services.
References to non-CDC sites on the Internet are
provided as a service to MMWR readers and do not constitute or imply
endorsement of these organizations or their programs by CDC or the U.S.
Department of Health and Human Services. CDC is not responsible for the content
of pages found at these sites. URL addresses listed in MMWR were current as of
the date of publication.
All HTML versions of MMWR articles are generated from final proofs through an automated process. This conversion might result in character translation or format errors in the HTML version. Users are referred to the electronic PDF version (https://www.cdc.gov/mmwr) and/or the original MMWR paper copy for printable versions of official text, figures, and tables.
Questions or messages regarding errors in formatting should be addressed to mmwrq@cdc.gov.
- Page last reviewed: August 14, 2017
- Page last updated: August 14, 2017
- Content source:


 ShareCompartir
ShareCompartir