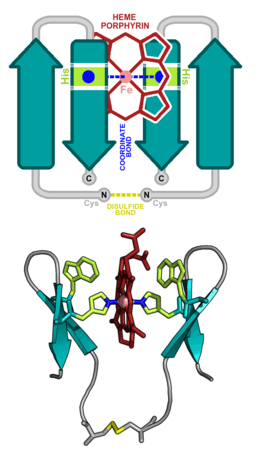
Hemeprotein
A hemeprotein (or haemprotein; also hemoprotein or haemoprotein), or heme protein, is a protein that contains a heme prosthetic group. They are very large class of metalloproteins. The heme group confers functionality, which can include oxygen carrying, oxygen reduction, electron transfer, and other processes. Heme is bound to the protein either covalently or noncovalently or both.[1]
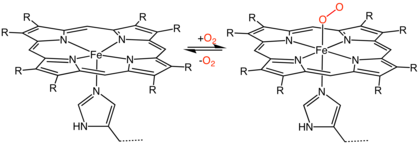
The heme consists of iron cation bound at the center of the conjugate base of the porphyrin, as well as other ligands attached to the "axial sites" of the iron. The porphyrin ring is a planar dianionic, tetradentate ligand. The iron is typically Fe2+ or Fe3+. One or two ligands are attached at the axial sites. The porphyrin ring has 4 nitrogen atoms that bind to the iron, leaving two other coordination positions of the iron available for bonding to the histidine of the protein and a divalent atom.[1]
Hemeproteins probably evolved to incorporate the iron atom contained within the protoporphyrin IX ring of heme into proteins. As it makes hemeproteins responsive to molecules that can bind divalent iron, this strategy has been maintained throughout evolution as it plays crucial physiological functions. Oxygen (O2), nitric oxide (NO), carbon monoxide (CO) and hydrogen sulfide (H2S) bind to the iron atom in heme proteins. Once bound to the prosthetic heme groups, these molecules can modulate the activity/function of those hemeproteins, affording signal transduction. Therefore, when produced in biologic systems (cells), these gaseous molecules are referred to as gasotransmitters.
Because of their diverse biological functions and widespread abundance, hemeproteins are among the most studied biomolecules.[2] Data on heme protein structure and function has been aggregated into The Heme Protein Database (HPD), a secondary database to the Protein Data Bank.[3]
Roles
- Hemeproteins have diverse biological functions including oxygen transport, which is completed via hemeproteins including hemoglobin, myoglobin, neuroglobin, cytoglobin, and leghemoglobin.[4]
- Some hemeproteins - cytochrome P450s, cytochrome c oxidase, ligninases, catalase. and peroxidases - are enzymes. They often activate O2 for oxidation or hydroxylation.
- Hemeproteins also enable electron transfer as they form part of the electron transport chain. Cyctochrome a, cytochrome b, and cytochrome c have such electron transfer functions.
The sensory system also relies on some hemeproteins including FixL, an oxygen sensor, CooA, a carbon monoxide sensor, and soluble guanylyl cyclase.
Hemoglobin and myoglobin
Hemoglobin and myoglobin are examples of hemeproteins that respectively transport and store of oxygen in mammals. Hemoglobin is a quaternary protein that occurs in the red blood cell, whereas, myoglobin is a tertiary protein found the muscle cells of mammals. Although they might differ in location and size, their function are similar. Being hemeproteins, they both contain a heme prosthetic group.
His-F8 of the myoglobin, also known as the proximal histidine, is covalently bonded to the 5th coordination position of the iron. Oxygen interacts with the distal His by way of a hydrogen bond, not a covalent one. It binds to the 6th coordination position of the iron, His-E7 of the myoglobin binds to the oxygen that is now covalently bonded to the iron. The same is true for hemoglobin; however, being a protein with four subunits, hemoglobin contains four heme units in total, allowing four oxygen molecules in total to bind to the protein.
Myoglobin and hemoglobin are globular proteins that serve to bind and deliver oxygen. These globins dramatically improve the concentration of molecular oxygen that can be carried in the biological fluids of vertebrates and some invertebrates.
Both of these globins contain an virginal prosthetic group for binding oxygen and they contain many similar structural characteristics. Yet the differences in function between these two proteins can be explored from a structural perspective, leading to a discourse on ligand binding and allosteric regulation.
Myoglobin and hemoglobin are of obvious critical importance to vertebrate life, yet they are also studied because they serve as well-characterized models for illustrating the principles of protein structure, dynamics, and function.
Myoglobin
Myoglobin is found in vertebrate muscle cells. Muscle cells, when put into action, can quickly require a large amount of oxygen for respiration because of their incredible demand for energy. Therefore, muscle cells use myoglobin to accelerate oxygen diffusion and act as localized oxygen reserves for times of intense respiration. myoglobin also stores the required amount of oxygen and make it available for the skeletal muscle cell mitochondria
Hemoglobin
In vertebrates, hemoglobin is found in the cytosol of red blood cells in the bloodstream. Hemoglobin is sometimes referred to as the oxygen transport protein, in order to contrast it with its stationary cousin myoglobin, although its function and mechanism are more complex than this name would suggest.
In vertebrates, oxygen is taken into the body by the tissues of the lungs, and passed to the red blood cells in the bloodstream. Oxygen is then distributed to all of the tissues in the body and offloaded from the red blood cells to respiring cells. Hemoglobin then picks up carbon dioxide to be returned to the lungs. Thus, hemoglobin binds and offloads both oxygen and carbon dioxide at the appropriate tissues, serving to deliver the oxygen needed for cellular metabolism and removing the resulting waste product, CO2.
Cytochrome c oxidase
Cytochrome c oxidase is an enzyme embedded in the inner membrane of mitochondria. Its main function is to oxidise the Cytochrome c protein. Cytochrome c oxidase contains several metal active sites.
Designed heme proteins
Due to the diverse functions of the heme molecule: as an electron transporter, an oxygen carrier, and as an enzyme cofactor, heme binding proteins have consistently attracted the attention of protein designers. Initial design attempts focused on α-helical heme binding proteins, in part, due to the relative simplicity of designing self-assembling helical bundles. Heme binding sites were designed inside the inter-helical hydrophobic grooves. Examples of such designs include:
Later design attempts focused on creating functional heme binding helical bundles, such as:
- Oxidoreductases
- Peroxidases
- Electron transport proteins
- Oxygen transport proteins
- Photosensitive proteins
Design techniques have matured to such an extent that it is now possible to generate entire libraries of heme binding helical proteins .
Recent design attempts have focused on creating all-beta heme binding proteins, whose novel topology is very rare in nature. Such designs include:
References
- Nelson, D. L.; Cox, M. M. "Lehninger, Principles of Biochemistry" 3rd Ed. Worth Publishing: New York, 2000. ISBN 1-57259-153-6.
- Gibney, Brian R.; Elvekrog, M. M.; Reedy, C. J. (18 September 2007). "Development of a heme protein structure electrochemical function database". Nucleic Acids Research. 36 (Database): D307–D313. doi:10.1093/nar/gkm814. PMC 2238922. PMID 17933771.
- Heme Protein Database
- S. J. Lippard, J. M. Berg “Principles of Bioinorganic Chemistry” University Science Books: Mill Valley, CA; 1994. ISBN 0-935702-73-3.
External links
- Heme Protein Database
- Hemeproteins at the US National Library of Medicine Medical Subject Headings (MeSH)