Gram stain
Gram stain or Gram staining, also called Gram's method, is a method of staining used to distinguish and classify bacterial species into two large groups (Gram-positive and Gram-negative). The name comes from the Danish bacteriologist Hans Christian Gram, who developed the technique.[1]
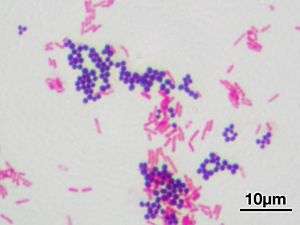
Gram staining differentiates bacteria by the chemical and physical properties of their cell walls. Gram-positive cells have a thick layer of peptidoglycan in the cell wall that retains the primary stain, crystal violet. Gram-negative cells have a thinner peptidoglycan layer that allows the crystal violet to wash out. They are stained pink by the counterstain,[2] commonly safranin or fuchsine.
The Gram stain is almost always the first step in the preliminary identification of a bacterial organism. While Gram staining is a valuable diagnostic tool in both clinical and research settings, not all bacteria can be definitively classified by this technique. This gives rise to Gram-variable and Gram-indeterminate groups.
History
The method is named after its inventor, the Danish scientist Hans Christian Gram (1853–1938), who developed the technique while working with Carl Friedländer in the morgue of the city hospital in Berlin in 1884. Gram devised his technique not for the purpose of distinguishing one type of bacterium from another but to make bacteria more visible in stained sections of lung tissue.[3] He published his method in 1884, and included in his short report the observation that the typhus bacillus did not retain the stain.[4]
Uses
Gram staining is a bacteriological laboratory technique[5] used to differentiate bacterial species into two large groups (Gram-positive and Gram-negative) based on the physical properties of their cell walls.[6] Gram staining is not used to classify archaea, formerly archaeabacteria, since these microorganisms yield widely varying responses that do not follow their phylogenetic groups.[7]
The Gram stain is not an infallible tool for diagnosis, identification, or phylogeny, and it is of extremely limited use in environmental microbiology. It is used mainly to make a preliminary morphologic identification or to establish that there are significant numbers of bacteria in a clinical specimen. It cannot identify bacteria to the species level, and for most medical conditions, it should not be used as the sole method of bacterial identification. In clinical microbiology laboratories, it is used in combination with other traditional and molecular techniques to identify bacteria. Some organisms are Gram-variable (meaning they may stain either negative or positive); some are not stained with either dye used in the Gram technique and are not seen. In a modern environmental or molecular microbiology lab, most identification is done using genetic sequences and other molecular techniques, which are far more specific and informative than differential staining.
Gram staining has been suggested to be as effective a diagnostic tool as PCR in one primary research report regarding gonococcal urethritis.[8]
Medical
Gram stains are performed on body fluid or biopsy when infection is suspected. Gram stains yield results much more quickly than culturing, and is especially important when infection would make an important difference in the patient's treatment and prognosis; examples are cerebrospinal fluid for meningitis and synovial fluid for septic arthritis.[5][9]
Staining mechanism

Gram-positive bacteria have a thick mesh-like cell wall made of peptidoglycan (50–90% of cell envelope), and as a result are stained purple by crystal violet, whereas Gram-negative bacteria have a thinner layer (10% of cell envelope), so do not retain the purple stain and are counter-stained pink by safranin. There are four basic steps of the Gram stain:
- Applying a primary stain (crystal violet) to a heat-fixed smear of a bacterial culture. Heat fixation kills some bacteria but is mostly used to affix the bacteria to the slide so that they don't rinse out during the staining procedure.
- The addition of iodide, which binds to crystal violet and traps it in the cell
- Rapid decolorization with ethanol or acetone
- Counterstaining with safranin.[10] Carbol fuchsin is sometimes substituted for safranin since it more intensely stains anaerobic bacteria, but it is less commonly used as a counterstain.[11]
| Application of | Reagent | Cell color | |||
|---|---|---|---|---|---|
| Gram-positive | Gram-negative | ||||
| Primary dye | crystal violet | purple | purple | ||
| Trapping agent | iodine | purple | purple | ||
| Decolorizer | alcohol/acetone | purple | colorless | ||
| Counter stain | safranin/carbol fuchsin | purple | pink | ||
Crystal violet (CV) dissociates in aqueous solutions into CV+
and chloride (Cl−
) ions. These ions penetrate the cell wall of both Gram-positive and Gram-negative cells. The CV+
ion interacts with negatively charged components of bacterial cells and stains the cells purple.[12]
Iodide (I−
or I−
3) interacts with CV+
and forms large complexes of crystal violet and iodine (CV–I) within the inner and outer layers of the cell. Iodine is often referred to as a mordant, but is a trapping agent that prevents the removal of the CV–I complex and, therefore, colors the cell.[13]
When a decolorizer such as alcohol or acetone is added, it interacts with the lipids of the cell membrane.[14] A Gram-negative cell loses its outer lipopolysaccharide membrane, and the inner peptidoglycan layer is left exposed. The CV–I complexes are washed from the gram-negative cell along with the outer membrane.[15] In contrast, a Gram-positive cell becomes dehydrated from an ethanol treatment. The large CV–I complexes become trapped within the Gram-positive cell due to the multilayered nature of its peptidoglycan.[15] The decolorization step is critical and must be timed correctly; the crystal violet stain is removed from both Gram-positive and negative cells if the decolorizing agent is left on too long (a matter of seconds).[16]
After decolorization, the Gram-positive cell remains purple and the Gram-negative cell loses its purple color.[16] Counterstain, which is usually positively charged safranin or basic fuchsine, is applied last to give decolorized Gram-negative bacteria a pink or red color.[2][17] Both Gram-positive bacteria and Gram-negative bacteria pick up the counterstain. The counterstain, however, is unseen on Gram-positive bacteria because of the darker crystal violet stain.
Examples
Gram-positive bacteria
Gram-positive bacteria generally have a single membrane (monoderm) surrounded by a thick peptidoglycan. This rule is followed by two phyla: Firmicutes (except for the classes Mollicutes and Negativicutes) and the Actinobacteria.[6][18] In contrast, members of the Chloroflexi (green non-sulfur bacteria) are monoderms but possess a thin or absent (class Dehalococcoidetes) peptidoglycan and can stain negative, positive or indeterminate; members of the Deinococcus–Thermus group stain positive but are diderms with a thick peptidoglycan.[6][18]
Historically, the Gram-positive forms made up the phylum Firmicutes, a name now used for the largest group. It includes many well-known genera such as Lactobacillus, Bacillus, Listeria, Staphylococcus, Streptococcus, Enterococcus, and Clostridium.[19] It has also been expanded to include the Mollicutes, bacteria like Mycoplasma, Thermoplasma that lack cell walls and so cannot be Gram-stained, but are derived from such forms.[20]
Some bacteria have cell walls which are particularly adept at retaining stains. These will appear positive by Gram stain even though they are not closely related to other Gram-positive bacteria. These are called acid-fast bacteria, and can only be differentiated from other Gram-positive bacteria by special staining procedures.[21]
Gram-negative bacteria
Gram-negative bacteria generally possess a thin layer of peptidoglycan between two membranes (diderms). Most bacterial phyla are Gram-negative, including the cyanobacteria, green sulfur bacteria, and most Proteobacteria (exceptions being some members of the Rickettsiales and the insect-endosymbionts of the Enterobacteriales).[6][18]
Gram-variable and Gram-indeterminate bacteria
Some bacteria, after staining with the Gram stain, yield a Gram-variable pattern: a mix of pink and purple cells are seen.[15] In cultures of Bacillus, Butyrivibrio, and Clostridium, a decrease in peptidoglycan thickness during growth coincides with an increase in the number of cells that stain Gram-negative.[22] In addition, in all bacteria stained using the Gram stain, the age of the culture may influence the results of the stain.[22]
Gram-indeterminate bacteria do not respond predictably to Gram staining and, therefore, cannot be determined as either Gram-positive or Gram-negative. Examples include many species of Mycobacterium, including Mycobacterium bovis, Mycobactrium leprae and Mycobacterium tuberculosis, the latter two of which are the causative agents of leprosy and tuberculosis respectively.[23][24]
Orthographic note
The term "Gram staining" is derived from the surname of Hans Christian Gram; the eponym (Gram) is therefore capitalized but not the common noun (stain) as is usual for scientific terms.[25] The initial letters of "Gram-positive" and "Gram-negative", which are eponymous adjectives, can be either lowercase "g" or capital "G", depending on what style guide (if any) governs the document being written. Lowercase style is used by the US Centers for Disease Control and Prevention and other style regimens such as the AMA style.[26] Dictionaries may use lowercase,[27][28] uppercase,[29][30][31][32] or both.[33][34] Uppercase 'Gram-positive' or 'Gram-negative' usage is also common in many scientific journal articles and publications.[34][35][36] When articles are submitted to journals, each journal may or may not apply house style to the postprint version. Preprint versions contain whichever style the author happened to use. Even style regimens that use lowercase for the adjectives "gram-positive" and "gram-negative" still use capital for "Gram stain".
See also
- Bacterial cell structure
- Ziehl–Neelsen stain
References
- Colco, R (2005). "Gram Staining". Current Protocols in Microbiology. 00 (1): Appendix 3C. doi:10.1002/9780471729259.mca03cs00. ISBN 978-0471729259. PMID 18770544.
- Beveridge T. J.; Davies J. A. (November 1983). "Cellular responses of Bacillus subtilis and Escherichia coli to the Gram stain". Journal of Bacteriology. 156 (2): 846–58. PMC 217903. PMID 6195148.
- Austrian, R. (1960). "The Gram stain and the etiology of lobar pneumonia, an historical note". Bacteriological Reviews. 24 (3): 261–265. PMC 441053. PMID 13685217.
- Gram, H.C. (1884). "Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten". Fortschritte der Medizin (in German). 2: 185–189..
English translation in: Brock, T.D. (1999). Milestones in Microbiology 1546–1940 (2 ed.). ASM Press. pp. 215–218. ISBN 978-1-55581-142-6.
Translation is also at: Brock, T.D. "Pioneers in Medical Laboratory Science: Christian Gram 1884". Hoslink. Retrieved 2010-07-27. Cite journal requires|journal=(help) - Ryan K.J., Ray C.G. (editors) (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. pp. 232f. ISBN 978-0838585290.CS1 maint: extra text: authors list (link)
- Madigan, M.T.; Martinko J.; Parker J. (2004). Brock Biology of Microorganisms (10th ed.). Lippincott Williams & Wilkins. ISBN 978-0-13-066271-2.
- Beveridge T.J. (2001). "Use of the Gram stain in microbiology". Biotechnic & Histochemistry. 76 (3): 111–118. doi:10.1080/714028139. PMID 11475313.
- El-Garnal, A.H., Al-Otaibi, S.R., Alshamali, A., Abdulrazzaq, A., Najem, N., and Fouzan, A.A. Polymerase chain reaction is no better than Gram stain for diagnosis of gonococcal urethritis. Indian Journal of Dermatology, Venereology and Leprology, (2009); 75, 101.
- Søgaard M.; Nørgaard M.; Schønheyder H. (2007). "First notification of positive blood cultures: high accuracy of the Gram stain report". Journal of Clinical Microbiology. 45 (4): 1113–1117. doi:10.1128/JCM.02523-06. PMC 1865800. PMID 17301283.
- Microbiology: Principles and Explorations, p 65; Jacquelyn G. Black, Prentice Hall, 1993.
- "Medical Chemical Corporation". med-chem.com. Retrieved 9 March 2016.
- Leboffe, Michael (2014). Microbiology Laboratory Theory and Application (3rd ed.). Englewood, CO: Morton Publishing Company. p. 105. ISBN 978-1617312809.
- "StainsFile - Stain theory - What a mordant is not". stainsfile.info. Retrieved 9 March 2016.
- "Welcome to Microbugz - Gram Stain". www.austincc.edu. Retrieved 2017-05-26.
- Tim, Sandle (2015-10-21). Pharmaceutical microbiology : essentials for quality assurance and quality control. ISBN 9780081000229. OCLC 923807961.
- Hardy, Jay; Maria, Santa. "Gram's Serendipitous Stain" (PDF). Hardy's Diagnostics.
- Davies J. A.; Anderson G. K.; Beveridge T. J.; Clark H. C. (November 1983). "Chemical mechanism of the Gram stain and synthesis of a new electron-opaque marker for electron microscopy, which replaces the iodine mordant of the stain". Journal of Bacteriology. 156 (2): 837–845. PMC 217902. PMID 6195147.
- Don J. Brenner, Noel R. Krieg, James T. Staley (July 26, 2005) [1984]. George M. Garrity (ed.). Introductory Essays. Bergey's Manual of Systematic Bacteriology. 2A (2nd ed.). New York: Springer. p. 304. ISBN 978-0-387-24143-2. British Library no. GBA561951.CS1 maint: uses authors parameter (link)
- Galperin, Michael Y. (2013-12-27). "Genome Diversity of Spore-Forming Firmicutes". Microbiology Spectrum. 1 (2). doi:10.1128/microbiolspectrum.tbs-0015-2012. ISSN 2165-0497. PMC 4306282. PMID 26184964.
- Practical Medical Microbiology by Hams H. Hashem, from http://qu.edu.iq/el/mod/resource/view.php?id=1391
- "The Acid Fast Stain". www2.highlands.edu. Retrieved 2017-06-09.
- Beveridge, Terry J. (March 1990). "Mechanism of gram variability in select bacteria". Journal of Bacteriology. 172 (3): 1609–20. doi:10.1128/jb.172.3.1609-1620.1990. PMC 208639. PMID 1689718.
- Black, Jacquelyn (2012). Microbiology: Principles and Exploration (8th ed.). John Wiley & Sons. p. 68. ISBN 978-0-470-54109-8.
- Reynolds J.; Moyes R.B.; Breakwell D.P. (2009). "Differential staining of bacteria: acid fast stain". Current Protocols in Microbiology. Appendix 3: Appendix 3H. doi:10.1002/9780471729259.mca03hs15. ISBN 978-0471729259. PMID 19885935.
- Waddingham, Anne (28 August 2014). New Hart's Rules: The Oxford Style Guide. OUP Oxford. p. 105. ISBN 978-0199570027.
- Centers for Disease Control and Prevention. Emerging Infectious Diseases Journal Style Guide. Preferred Usage
- Elsevier, Dorland's Illustrated Medical Dictionary, Elsevier.
- Merriam-Webster, gram–positive, Merriam-Webster.
- "Definition of Gram-positive". Collins.
- "Gram stain". Oxford Dictionary.
- "Definition of Gram-positive". Medicinenet.
- "Gram negative/positive". Business dictionary.
- "gram-pos·i·tive or Gram-pos·i·tive". The American Heritage Dictionary.
- "Gram-positive". Dictionary.com.
- Lisa Brown, Julie M. Wolf, Rafael Prados-Rosales & Arturo Casadevall (2015). "Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi". Nature Reviews Microbiology. 13 (10): 620–630. doi:10.1038/nrmicro3480. PMC 4860279. PMID 26324094.CS1 maint: uses authors parameter (link)
- Kristen L. Mueller (12 June 2015). "Detecting Gram-negative bacteria". Science. 348 (6240): 1218. doi:10.1126/science.348.6240.1218-o.
External links
| Wikimedia Commons has media related to Gram stains. |
| The Wikibook School Science has a page on the topic of: Gram staining |