Questions and Answers about Lab Testing

General Laboratory Testing
Questions in This Section
Q: What specimens should be collected from patients who meet the mumps clinical case definition?
A: CDC recommends that a buccal or oral swab specimen and a blood specimen be collected from all patients with clinical features compatible with mumps. See Specimen Collection, Storage, and Shipment for detailed instructions regarding recommended samples for mumps testing.
Q: When is the optimal timing for collection of specimens for laboratory confirmation of mumps?
A: The acute-phase serum and clinical samples for detection of virus should be collected as soon as possible upon suspicion of mumps disease. The early collection of buccal swab specimens provides the best means of laboratory confirmation, particularly among suspected mumps patients with a history of vaccination (Rota et al, 2013). If the acute-phase serum sample collected ≤3 days after parotitis onset is negative, and the case has a negative (or not done) result for RT-PCR, a second serum sample collected 5–10 days after symptom onset is recommended because, in some cases, the IgM response is not detectable until 5 days after symptom onset. In an outbreak setting (see Figure below), the proportion of samples with positive results by RT-PCR (N target gene) decreased as the interval between onset and specimen collection increased (Rota et al., 2013). The trend was reversed for IgM detection with increasing time to sample collection the number of seropositive samples increased.
Percentage of Mumps Specimens Determined Positive by CDC IgM Capture EIA or rRT-PCR (N target gene) as a Function of Time Post Parotitis Onset*
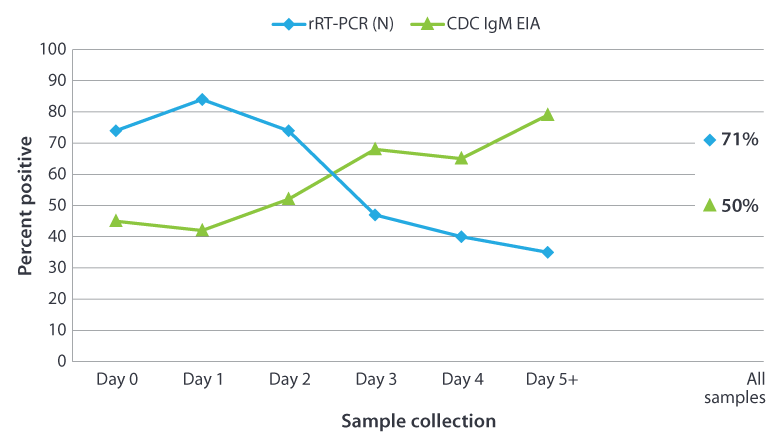
The percentage of positive results obtained from testing 296 confirmed mumps cases from New York City by day of sample collection after onset of symptoms. The serum samples were tested for presence of IgM using the CDC capture IgMEIA. The buccal swab samples were tested by rRT-PCR using the mumps nucleoprotein (N) gene as the target.
*Done in collaboration with New York City Deparment of Health and Mental Hygiene Public Health Laboratory, New York, NY
Mumps virus was isolated from 209 (71%) of the 296 buccal swabs tested.
Q: What tests are available for laboratory confirmation of mumps?
A: Standard serologic assays that detect mumps IgM are commercially available in both EIA and IFA formats. Viral detection methods include standard methods of culturing virus in appropriate cell lines and techniques, such as real time RT-PCR to detect mumps viral RNA. RT-PCR is available at many state public health laboratories and though the APHL/CDC Vaccine Preventable Disease Reference Centers (VPD-RC [2 pages]).
Q: Where do I send laboratory specimens for testing?
Detection of Mumps Virus in Clinical Samples
Questions in This Section
- Why should I ask for buccal or oral swab specimens in addition to the serum sample?
- What does a mumps-negative RT-PCR result mean?
- What does a mumps-positive RT-PCR result mean?
- Why attempt to isolate mumps virus in cell culture?
- What is a mumps genotype?
- What mumps virus genotype has been commonly detected in the United States?
- Does mumps vaccine protect against commonly detected mumps virus genotypes in the United States?
- Are there small genetic changes that may be affecting our molecular diagnostics?
- Would genetic changes that are occurring cause the vaccine not to work against certain strains of the virus?
- What is the gold standard for laboratory confirmation of mumps?
- If a clinical sample is determined to be positive by RT-PCR or by viral isolation at the state laboratory, is it necessary to send a sample to CDC for sequence analysis?
- Where can I find RT-PCR and real-time RT-PCR protocols for mumps?
- Where do I obtain material for RT-PCR and virus isolation controls?
Q: Why should I ask for buccal or oral swab specimens in addition to the serum sample?
A: Buccal and oral swab samples enhance the ability to laboratory-confirm a mumps infection and can provide additional information (sequence analysis to obtain the mumps genotype) to aid epidemiologic investigations. Because serologic tests cannot differentiate between an exposure to vaccine and an exposure to wild-type mumps virus, it is necessary to obtain other clinical samples to determine the genotype (wild-type vs. vaccine). Mumps infection can also be confirmed by detection of mumps specific IgM in serum samples.
Q: What does a mumps-negative RT-PCR result mean?
A: Failure to detect mumps virus RNA by RT-PCR in samples from a person with clinically compatible mumps symptoms does not rule out mumps as a diagnosis. Successful detection of mumps virus depends primarily on the timing of collection and quality of the clinical sample. Vaccinated individuals may shed virus for a shorter period and might shed smaller amounts of virus, thus degradation of the sample has greater consequences for successful detection of virus. In outbreaks among two-dose vaccine recipients, mumps virus RNA was detected in samples from 30%–71% of case-patients if the samples were collected within 3 days following onset of parotitis. IgM was detected in 13 to 50% of these cases (Bitsko et al, 2008, Rota et al. 2009 Rota et al.2013).
Q: What does a mumps-positive RT-PCR result mean?
A: A positive RT-PCR signal indicates the presence of mumps virus RNA in the patient sample. The positive result should be used only to support a clinical diagnosis of mumps. A positive RT-PCR result provides laboratory confirmation of mumps infection in persons with symptoms consistent with mumps who have not been vaccinated within the preceding 45 days.
Q: Why attempt to isolate mumps virus in cell culture?
A: Virus isolation is considered among the best methods for confirming mumps infection. Virus can be detected when IgM antibodies or a rise in IgG titer are not detected. Often it is necessary to grow the virus in culture to have adequate material for viral sequencing. Sequence analysis allows the determination of the mumps genotype (there are currently 12 recognized genotypes). The sequence information can help to identify the source of the virus and can provide confirmation of suspected epidemiologic links. In addition, virus isolation is less likely than PCR assays to give false-positive results due to contamination.
Q: What is a mumps genotype?
A: Mumps strains are assigned to 1 of 12 genotypes based on the sequences of the gene coding for the short hydrophobic (SH) protein. In some circumstances, a genotype has been associated with endemic circulation of mumps virus in a country; however, routine genotype surveillance for mumps is limited to only a few countries. The genetic information from circulating mumps viruses is used to track the transmission pathways of the virus and can be used to suggest epidemiologic links, or lack thereof, between cases and outbreaks.
1Jin, L., et al., Genomic diversity of mumps virus and global distribution of the 12 genotypes. Rev Med Virol, 2015. 25(2): p. 85-101
Q: What mumps virus genotype has been commonly detected in the United States?
A: CDC has detected mostly genotype G among people with mumps in the United States since we initiated routine genotype surveillance for mumps in 2006. We’ve also detected a few of the other 11 genotypes, but they are usually associated with mumps importations into the United States, and have not been associated with large outbreaks.
Mumps outbreaks are typically associated with only one genotype. There are no differences in the genotypes detected in vaccinated and unvaccinated people who have gotten mumps in the United States.
Q: Does mumps vaccine protect against commonly detected mumps virus genotypes in the United States?
A: Yes. The mumps vaccine protects against the mumps virus genotype G, the most common type detected in the United States. A lab study showed that during a large U.S. outbreak in 2006, mumps vaccine (genotype A Jeryl-Lynn mumps virus vaccine strain) produced antibodies that effectively fought the genotype G mumps virus.
For more information, see the scientific article about the study.
Q: Are there small genetic changes that may be affecting our molecular diagnostics?
A: Viruses like mumps have very high rates of genetic change compared to other organisms. Our diagnostic assay, RT-PCR, is designed to detect a specific sequence that is conserved in many mumps genotypes. If genetic changes occur in the sequence that is being detected by RT-PCR, the assay may lose the ability to detect mumps virus with high sensitivity. CDC monitors the performance of the RT-PCR assay in collaboration with the four Vaccine Preventable Disease Reference Centers that are supported by the Association of Public Health Laboratories (APHL) and CDC. In addition, state laboratories performing the RT-PCR assay to detect mumps may participate in a proficiency testing program that was developed by CDC and the Reference Centers and is managed by the Wisconsin State Laboratory of Hygiene.
Q: Would genetic changes that are occurring cause the vaccine not to work against certain strains of the virus?
A: Not all genetic changes result in a change in viral proteins. "Silent mutations" change the genetic sequence without changing the actual protein. The viral proteins are recognized by the host’s immune system. There several viral proteins, each containing many recognition sites. Therefore, it would take many genetic changes to change the viral proteins to the point that they are no longer recognized by the immune response in vaccinated individuals. CDC is developing tests to monitor the ability of serum from vaccinated individuals to neutralize currently circulating strains.
Q: What is the gold standard for laboratory confirmation of mumps?
A: Virus culture is the gold standard for mumps confirmation. However, sample quality must be maintained to ensure viability of the virus. Laboratories are strongly encouraged to perform cell culture isolation of mumps from buccal or oral swab specimens. Primary monkey kidney cells and Vero cells are frequently used to isolate virus. Confirmation of successful isolation of mumps can be performed using immunofluorescent antibody staining or standard RT-PCR. Unfortunately, virus isolation can require several days to several weeks to complete, while detection IgM by EIA or viral RNA by RT-PCR can usually be performed in less than one day.
Q: If a clinical sample is determined to be positive by RT-PCR or by viral isolation at the state laboratory, is it necessary to send a sample to CDC for sequence analysis?
A: Laboratories are encouraged to send patient samples from positive sporadic cases of mumps as well as representative samples from an outbreak. The sequence of the mumps short hydrophobic (SH) gene is used to assign mumps viruses to one of 12 recognized genotypes. The sequence information will help to identify the source of the virus and can provide confirmation of suspected epidemiologic links. Samples for genotyping can be sent to the CDC or the VPD-RCs.
Q: Where can I find RT-PCR and real-time RT-PCR protocols for mumps?
A: The following protocols are available online:
- Real-time RT-PCR protocol, targeting the nucleoprotein (N) gene: Word [8 pages] | PDF [8 pages] 508 compliant
- Standard RT-PCR for mumps virus: Word [6 pages] | PDF [6 pages] 508 compliant
Q: Where do I obtain material for RT-PCR and virus isolation controls?
A: CDC can provide a sample of RNA that has been purified from cells infected with mumps virus. This material is ready to use in RT-PCR reactions. If laboratories would like to produce their own RNA samples or require a positive control for virus isolation, CDC can provide a sample of wild-type mumps virus. Public health laboratories or laboratories affiliated with state public health laboratories may send request for mumps RNA or virus to prota@cdc.gov. A proficiency test for mumps RT-PCR can be obtained from the Wisconsin State Laboratory of Hygiene.
References
- Bitsko RH, Cortese MM, Dayan GH, Rota PA, Lowe L, Iversen SC, Bellini WJ. Detection of RNA of mumps virus during an outbreak in a population with a high level of measles, mumps, and rubella vaccine coverage. J Clin Microbiol 2008;46:1101–3.
- Briss PA, Fehrs LJ, Parker RA, Wright PF, Sannella EC, Hutcheson RH, Schaffner W. Sustained transmission of mumps in a highly vaccinated population: assessment of primary vaccine failure and waning vaccine-induced immunity. J Infect Dis 1994;169:77–82.
- Davidkin I, Jokinen S, Paananen A, Leinikki P, Peltola H. Etiology of mumps-like illnesses in children and adolescents vaccinated for measles, mumps, and rubella. J Infect Dis 3005;191:719–23.
- Gut JP, Lablache C, Behr S, Kirn A. Symptomatic mumps virus reinfections. J Med Virol 1995;45:17–23.
- Jin, L., et al., Genomic diversity of mumps virus and global distribution of the 12 genotypes. Rev Med Virol, 2015. 25(2): p. 85-101.
- Krause CH., Molyneaux PJ, Ho-Yen DO, McIntyre P, Carman WF, Templeton KE. Comparison of mumps-IgM ELISAs in acute infection. J Clin Virol 2007;38:153–6.
- Narita M, Matsuzono Y, Takekoshi Y, Yamada S, Itakura O, Kubota M, Kikuta H, Togashi T. Analysis of mumps vaccine failure by means of avidity testing for mumps virus-specific immunoglobulin G. Clin Diagn Lab Immunol 1998;5:799–803.
- Rota JS, Rose JB, Doll MK, McNall RJ, McGrew M, Williams N, Lopareva EN, Barskey AE, Punsalang Jr A, Rota PA, Oleszko WR, Hickman CJ, Zimmerman DM, Bellini WJ. Comparison of the sensitivity of laboratory diagnostic methods from a well-characterized outbreak of mumps in New York City in 2009. Clin Vaccine Immunol. 2013;20:391-6.
- Rota JS, Turner JC, Yost-Daljev MK, Freeman M, Toney DM, Meisel E, Williams N, Sowers SB, Lowe L, Rota PA, Nicolai LA, Peake L, Bellini WJ. Investigation of a mumps outbreak among university students with two measles-mumps-rubella (MMR) vaccinations, Virginia, September–December 2006. J Med Virol 2009;81:1819–25.
- Sakata H, Tsurudome M, Hishiyama M, Ito Y, Sugiura A. Enzyme-linked immunosorbent assay for mumps IgM antibody: comparison of IgM capture and indirect IgM assay. J Virol Methods 1985;12:303–11.
- Sartorius B, Penttinen P, Nilsson J, Johansen K, Jönsson K, Arneborn M, Löfdahl M, Giesecke J. An outbreak of mumps in Sweden, February–April 2004. Euro Surveill 2005;10 (9):pii=559. Available at http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=559
- Page last reviewed: November 17, 2016
- Page last updated: February 14, 2017
- Content source:


 ShareCompartir
ShareCompartir