Lactic acid bacteria
Lactic acid bacteria (LAB) are an order of gram-positive, low-GC, acid-tolerant, generally nonsporulating, nonrespiring, either rod-shaped (bacilli) or spherical (cocci) bacteria that share common metabolic and physiological characteristics. These bacteria, usually found in decomposing plants and milk products, produce lactic acid as the major metabolic end product of carbohydrate fermentation. This trait has, throughout history, linked LAB with food fermentations, as acidification inhibits the growth of spoilage agents. Proteinaceous bacteriocins are produced by several LAB strains and provide an additional hurdle for spoilage and pathogenic microorganisms. Furthermore, lactic acid and other metabolic products contribute to the organoleptic and textural profile of a food item. The industrial importance of the LAB is further evidenced by their generally recognized as safe (GRAS) status, due to their ubiquitous appearance in food and their contribution to the healthy microbiota of animal and human mucosal surfaces. The genera that comprise the LAB are at its core Lactobacillus, Leuconostoc, Pediococcus, Lactococcus, and Streptococcus, as well as the more peripheral Aerococcus, Carnobacterium, Enterococcus, Oenococcus, Sporolactobacillus, Tetragenococcus, Vagococcus, and Weissella; these belong to the order Lactobacillales.
| Lactic acid bacteria | |
|---|---|
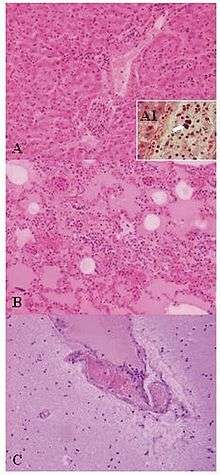 | |
| Lesions of Weissella confusa in the mona monkey (hematoxylin and eosin stain): A) liver: portal triads with neutrophilic infiltration (x10); A1, presence of bacterial emboli inside the vein (arrow) (x40). B) acute pneumonia: edema, congestion, and leukocyte cells exudation in the pulmonary alveoli (x10). C) encephalitis: congestion and marginalized neutrophils in nervous vessels (x10) | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Firmicutes |
| Class: | Bacilli |
| Order: | Lactobacillales |
| Families | |
| |
Characteristics
The lactic acid bacteria (LAB) are either rod-shaped (bacilli), or spherical (cocci), and are characterized by an increased tolerance to acidity (low pH range). This aspect helps LAB to outcompete other bacteria in a natural fermentation, as they can withstand the increased acidity from organic acid production (e.g., lactic acid). Laboratory media used for LAB typically include a carbohydrate source, as most species are incapable of respiration. LAB are catalase-negative. LAB are amongst the most important groups of microorganisms used in the food industry.[1] Their relative simple metabolism has also prompted their use as microbial cell factories for the production of several commodities for the food and non-food sectors [2]
Metabolism
Two main hexose fermentation pathways are used to classify LAB genera. Under conditions of excess glucose and limited oxygen, homolactic LAB catabolize one mole of glucose in the Embden-Meyerhof-Parnas pathway to yield two moles of pyruvate. Intracellular redox balance is maintained through the oxidation of NADH, concomitant with pyruvate reduction to lactic acid. This process yields two moles of ATP per mole of glucose consumed. Representative homolactic LAB genera include Lactococcus, Enterococcus, Streptococcus, Pediococcus, and group I lactobacilli [3]
Heterofermentative LAB use the pentose phosphate pathway, alternatively referred to as the pentose phosphoketolase pathway. One mole of glucose-6-phosphate is initially dehydrogenated to 6-phosphogluconate and subsequently decarboxylated to yield one mole of CO2. The resulting pentose-5-phosphate is cleaved into one mole glyceraldehyde phosphate (GAP) and one mole acetyl phosphate. GAP is further metabolized to lactate as in homofermentation, with the acetyl phosphate reduced to ethanol via acetyl-CoA and acetaldehyde intermediates. In theory, end products (including ATP) are produced in equimolar quantities from the catabolism of one mole of glucose. Obligate heterofermentative LAB include Leuconostoc, Oenococcus, Weissella, and group III lactobacilli [3]
Streptococcus reclassification
In 1985, members of the diverse genus Streptococcus were reclassified into Lactococcus, Enterococcus, Vagococcus, and Streptococcus based on biochemical characteristics, as well as molecular features. Formerly, streptococci were segregated primarily based on serology, which has proven to correlate well with the current taxonomic definitions. Lactococci (formerly Lancefield group N streptococci) are used extensively as fermentation starters in dairy production, with humans estimated to consume 1018 lactococci annually. Partly due to their industrial relevance, both L. lactis subspecies (L. l. lactis and L. l. cremoris) are widely used as generic LAB models for research. L. lactis ssp. cremoris, used in the production of hard cheeses, is represented by the laboratory strains LM0230 and MG1363. In similar manner, L. lactis ssp. lactis is employed in soft cheese fermentations, with the workhorse strain IL1403 ubiquitous in LAB research laboratories. In 2001, Bolotin et al. sequenced the genome of IL1403, which coincided with a significant shift of resources to understanding LAB genomics and related applications.
Phylogeny
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature[4] [5] and the phylogeny is based on 16S rRNA-based LTP release 106 by 'The All-Species Living Tree' Project.[6]
| Lactobacillales |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Lactobacillales part 2 (continued)
| Lactobacillales part 2 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Notes:
♠ Strains found at the National Center for Biotechnology Information, but not listed in the List of Prokaryotic names with Standing in Nomenclature
Bacteriophages
A broad number of food products, commodity chemicals, and biotechnology products are manufactured industrially by large-scale bacterial fermentation of various organic substrates. Because enormous amounts of bacteria are being cultivated each day in large fermentation vats, the risk that bacteriophage contamination rapidly brings fermentations to a halt and cause economical setbacks is a serious threat in these industries. The relationship between bacteriophages and their bacterial hosts is very important in the context of the food fermentation industry. Sources of phage contamination, measures to control their propagation and dissemination, and biotechnological defence strategies developed to restrain phages are of interest. The dairy fermentation industry has openly acknowledged the problem of phage contamination, and has been working with academia and starter culture companies to develop defence strategies and systems to curtail the propagation and evolution of phages for decades.[7]
Bacteriophage–host interaction
The first contact between an infecting phage and its bacterial host is the attachment of the phage to the host cell. This attachment is mediated by the phage's receptor binding protein (RBP), which recognizes and binds to a receptor on the bacterial surface. RBPs are also referred to as host-specificity protein, host determinant, and antireceptor. For simplicity, the RBP term will be used here. A variety of molecules have been suggested to act as host receptors for bacteriophages infecting LAB; among those are polysaccharides and (lipo)teichoic acids, as well as a single-membrane protein. A number of RBPs of LAB phages have been identified by the generation of hybrid phages with altered host ranges. These studies, however, also found additional phage proteins to be important for successful phage infection. Analysis of the crystal structure of several RBPs indicates that these proteins share a common tertiary folding, and support previous indications of the saccharide nature of the host receptor. Gram-positive LAB have a thick peptidoglycan layer, which must be traversed to inject the phage genome into the bacterial cytoplasm. Peptidoglycan-degrading enzymes are expected to facilitate this penetration, and such enzymes have been found as structural elements of a number of LAB phages.[7]
Probiotics
Probiotics are products aimed at delivering living, potentially beneficial, bacterial cells to the gut ecosystem of humans and other animals, whereas prebiotics are indigestible carbohydrates delivered in food to the large bowel to provide fermentable substrates for selected bacteria. Strains of LAB are the most common microbes employed as probiotics. Two principal kinds of probiotic bacteria, members of the genera Lactobacillus and Bifidobacterium, have been studied in detail.[1][8]
Most probiotic strains belong to the genus Lactobacillus. Probiotics have been evaluated in research studies in animals and humans with respect to antibiotic-associated diarrhoea, travellers' diarrhoea, pediatric diarrhoea, inflammatory bowel disease, and irritable bowel syndrome. In the future, probiotics possibly will be used for different gastrointestinal diseases, vaginosis, or as delivery systems for vaccines, immunoglobulins, and other therapies.[9]
Exopolysaccharides
The quest to find food ingredients with valuable bioactive properties has encouraged interest in exopolysaccharides from LAB. Functional food products that offer health and sensory benefits beyond their nutritional composition are becoming progressively more important to the food industry. The sensory benefits of exopolysaccharides are well established, and there is evidence for the health properties that are attributable to exopolysaccharides from LAB. However, there is a wide variation in molecular structures of exopolysaccharides and the complexity of the mechanisms by which physical changes in foods and bioactive effects are elicited.[10]
Lactic acid bacteria and dental plaque
LAB are able to synthesize levans from sucrose, and dextrans from glucose.[11] Glucans, such as dextran, enable bacteria to adhere to the surface of teeth, which in turn can cause tooth decay through the formation of dental plaque and production of lactic acid.[12] While the primary bacteria responsible for tooth decay is Streptococcus mutans, LAB are among the most common oral bacteria which cause dental cavities.[13]
Lactic acid bacteria and fermentation of beverages
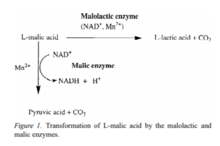
Lactic acid bacteria are used in the food industry for a variety of reasons such as the production of cheese and yogurt products. This process has been going on for thousands of years by human ancestors. But, some of the beverages we enjoy today are produced by using lactic acid bacteria. Popular drinks such as kombucha have known to have traces of lactobacillus and pediococcus once the drink is made.[14] Even the beer and wine-making process utilizes certain lactic acid bacteria mostly lactobacillus. The interesting relationship between lactic acid bacteria and yeast can be observed in the wine-making process. The LAB is used to start the wine-making process by starting the malolactic fermentation. After the malolactic fermentation, yeast cells are used to start the alcoholic fermentation process in grapes. The malolactic fermentation mechanism is mainly transformation of L-malic acid (dicarboxylic acid) to an lactic acid (monocarboxylic acid).[15] This change occurs due to the presence of malolactic and malic enzymes. All malic acid are degraded and this increase the pH levels which changes the taste of the wine.[15] Not only do they start the process but they are responsible for the different aromas produced in wine by the nutrients presence and the quality of the grapes. Also, the presence of different strains can change the desirability of aromas' presence. The different availability of enzymes that contribute to the vast spectrum of aromas in wine are associated with glycosidases, β-glucosidases, esterases, phenolic acid decarboxylases and citrate lyases.[16] By using molecular biology, researchers can help pick out different desirable strains that help improve the quality of wine and help with the removable of the undesirable strains. The same can be said about brewing beer as well which uses yeast with some breweries using lactic acid bacteria to change the taste of their beer.[17]
Lactic acid bacteria genera
See also
- Gaffkaemia
- Malolactic fermentation
- Lactic acid fermentation
- Lacto-2 RNA motif
References
- Sonomoto K, Yokota A, eds. (2011). Lactic Acid Bacteria and Bifidobacteria: Current Progress in Advanced Research. Caister Academic Press. ISBN 978-1-904455-82-0.
- Hatti-Kaul R, Chen L, Dishisha T, Enshasy HE (October 2018). "Lactic acid bacteria: from starter cultures to producers of chemicals". FEMS Microbiology Letters. 365 (20). doi:10.1093/femsle/fny213. PMID 30169778.
- Gänzle MG (2015). "Lactic metabolism revisited: metabolism of lactic acid bacteria in food fermentations and food spoilage". Current Opinion in Food Science. 2: 106–117. doi:10.1016/j.cofs.2015.03.001.
- See the List of Prokaryotic names with Standing in Nomenclature. Data extracted from Euzéby JP. "Lactobacillales". Archived from the original on 2013-01-27. Retrieved 2012-05-17.
- See the NCBI webpage on Lactobacillales Data extracted from "NCBI Taxonomy Browser". National Center for Biotechnology Information. Retrieved 2012-05-17.
- See 'The All-Species Living Tree' Project . Data extracted from the "16S rRNA-based LTP release 106 (full tree)" (PDF). Silva Comprehensive Ribosomal RNA Database. Retrieved 2012-05-17.
- Mc Grath S, van Sinderen D, eds. (2007). Bacteriophage: Genetics and Molecular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-14-1.
- Tannock G, ed. (2005). Probiotics and Prebiotics: Scientific Aspects (1st ed.). Caister Academic Press. ISBN 978-1-904455-01-1.
- Ljungh A, Wadstrom T, eds. (2009). Lactobacillus Molecular Biology: From Genomics to Probiotics. Caister Academic Press. ISBN 978-1-904455-41-7.
- Welman AD (2009). "Exploitation of Exopolysaccharides from lactic acid bacteria". Bacterial Polysaccharides: Current Innovations and Future Trends. Caister Academic Press. ISBN 978-1-904455-45-5.
- White D, Drummond J, Fuqua C (2012). The Physiology and Biochemistry of Prokaryotes (Fourth ed.). pp. 331–332. ISBN 978-0-19-539304-0.
- Brock biology of microorganisms (11th ed.). Pearson Prentice Hall. 2006. ISBN 978-0-13-144329-7.
- Tanzer JM, Livingston J, Thompson AM (October 2001). "The microbiology of primary dental caries in humans". Journal of Dental Education. 65 (10): 1028–37. PMID 11699974.
- Nguyen NK, Dong NT, Nguyen HT, Le PH (24 February 2015). "Lactic acid bacteria: promising supplements for enhancing the biological activities of kombucha". SpringerPlus. 4: 91. doi:10.1186/s40064-015-0872-3. PMC 4348356. PMID 25763303.
- Lonvaud-Funel A (1999). "Lactic acid bacteria in the quality improvement and depreciation of wine". Antonie van Leeuwenhoek. 76 (1–4): 317–31. doi:10.1023/A:1002088931106. PMID 10532386.
- Cappello MS, Zapparoli G, Logrieco A, Bartowsky EJ (February 2017). "Linking wine lactic acid bacteria diversity with wine aroma and flavour". International Journal of Food Microbiology. 243: 16–27. doi:10.1016/j.ijfoodmicro.2016.11.025. PMID 27940412.
- Dysvik A, Liland KH, Myhrer KS, Westereng B, Rukke E, de Rouck G, Wicklund T (2019). "Pre-fermentation with lactic acid bacteria in sour beer production". Journal of the Institute of Brewing. 125 (3): 342–356. doi:10.1002/jib.569.
Further reading
- Holzapfel WH, Wood BJ (1998). The genera of lactic acid bacteria (1st ed.). London Blackie Academic & Professional. ISBN 978-0-7514-0215-5.
- Salminen S, von Wright A, Ouwehand AC, eds. (2004). Lactic Acid Bacteria: Microbiological and Functional Aspects (3rd ed.). New York: Marcel Dekker, Inc. ISBN 978-0-8247-5332-0.
- Madigan MT, Martinko JM, Parker J (2004). Brock. Biología de los Microorganismos (10th ed.). Madrid: Pearson Educaciòn S.A. ISBN 978-84-205-3679-8.