Flagellin
Flagellin is a globular protein that arranges itself in a hollow cylinder to form the filament in a bacterial flagellum. It has a mass of about 30,000 to 60,000 daltons. Flagellin is the principal component of bacterial flagellum, and is present in large amounts on nearly all flagellated bacteria.
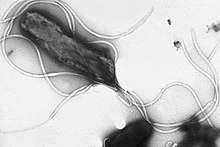
| Flagellin | |||||||
|---|---|---|---|---|---|---|---|
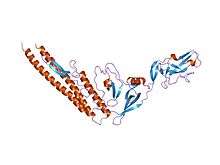 Homologous Salmonella flagellin, displaying the D0/D1 helical core as well as the D3 extension (PDB 1io1). | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | FliC | ||||||
| UniProt | P04949 | ||||||
| |||||||
| Flagellin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Flagellin | ||||||||
| Pfam | PF00700 | ||||||||
| InterPro | IPR001492 | ||||||||
| SCOPe | 1ucu / SUPFAM | ||||||||
| |||||||||
Structure
The structure of flagellin is responsible for the helical shape of the flagellar filament, which is important for its proper function.[1] It is transported through the center of the filament to the tip where it polymerases spontaneously into a part of the filament. It is unfolded by the FliS (P26608) flagellar secretion chaperone during transport.[2] The filament is made up of eleven smaller "protofilaments", nine of which contains flagellin in the L-type shape and the other two in the R-type shape.[3]
The helical N- and C-termini of flagellin form the inner core of the flagellin protein, and is responsible for flagellin's ability to polymerize into a filament. The middle residues make up the outer surface of the flagellar filament. While the termini of the protein are quite similar among all bacterial flagellins, the middle portion is wildly variable and can be absent in some species. The flagellin domains are numbered from the helical core (D0/D1) to the outside (D2, ...); when viewed from the amino-acid sequence, D0/D1 appears on the two termini. Flagellin-like structual proteins are found in other portions of the flagellum, such as the hook (flgE; P75937), the rod at the base, and the cap at the top.[4]
The middle part of E. coli (and related) flagellin, D3, displays a beta-folium fold and appears to maintain flagellar stability.[5]
Immune response
In mammals
Mammals often have acquired immune responses (T-cell and antibody responses)[6] to flagellated bacterium, which occur frequently to flagellar antigens. Flagellin has also been shown to directly interact with TLR5 on T cells.[7] Some bacteria are able to switch between multiple flagellin genes in order to evade this response.
The propensity of the immune response to flagellin may be explained by two facts:
- Flagellin is an extremely abundant protein in flagellated bacteria.
- There exists a specific innate immune receptor that recognizes flagellin, Toll-like receptor 5 (TLR5).[8]
In plants
In addition, a 22-amino acid sequence (flg22) of the conserved N-terminal part of flagellin is known to activate plant defence mechanisms.[9] Flagellin perception in Arabidopsis thaliana functions via the receptor-like-kinase FLS2 (FLAGELLIN SENSING 2).[10] Upon flg22 detection, FLS2 quickly binds to BAK1 (BRI1-associated kinase 1) to initiate signalling by reciprocal transphosphorylation of their kinase domains.[11] Mitogen-activated-protein-kinases (MAPK) acts as downstream signalling compounds, leading ultimately to PAMP-triggered immunity in which more than 900 genes are up-/down-regulated upon flg22 treatment.
Pre-stimulation with a synthetic flg22-peptide led to enhanced resistance against bacterial invaders.
References
- Steiner TS (February 2007). "How flagellin and toll-like receptor 5 contribute to enteric infection". Infection and Immunity. 75 (2): 545–52. doi:10.1128/IAI.01506-06. PMC 1828527. PMID 17118981.
- Vonderviszt F, Namba K (2013). Structure, Function and Assembly of Flagellar Axial Proteins. Austin, TX: Madame Curie Bioscience Database.
- Maki-Yonekura S, Yonekura K, Namba K (April 2010). "Conformational change of flagellin for polymorphic supercoiling of the flagellar filament". Nature Structural & Molecular Biology. 17 (4): 417–22. doi:10.1038/nsmb.1774. PMID 20228803.
- Imada K (April 2018). "Bacterial flagellar axial structure and its construction". Biophysical Reviews. 10 (2): 559–570. doi:10.1007/s12551-017-0378-z. PMC 5899737. PMID 29235079.
- Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K (March 2001). "Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling". Nature. 410 (6826): 331–7. Bibcode:2001Natur.410..331S. doi:10.1038/35066504. PMID 11268201.
- Genta RM (January 1997). "The immunobiology of Helicobacter pylori gastritis". Seminars in Gastrointestinal Disease. 8 (1): 2–11. PMID 9000497.
- Sharma N, Akhade AS, Qadri A (April 2013). "Sphingosine-1-phosphate suppresses TLR-induced CXCL8 secretion from human T cells". Journal of Leukocyte Biology. 93 (4): 521–8. doi:10.1189/jlb.0712328. PMID 23345392.
- Kathrani A, Holder A, Catchpole B, Alvarez L, Simpson K, Werling D, Allenspach K (2012). "TLR5 risk-associated haplotype for canine inflammatory bowel disease confers hyper-responsiveness to flagellin". PLOS ONE. 7 (1): e30117. Bibcode:2012PLoSO...730117K. doi:10.1371/journal.pone.0030117. PMC 3261174. PMID 22279566.
- García AV, Hirt H (2014-01-01). "Salmonella enterica induces and subverts the plant immune system". Frontiers in Microbiology. 5: 141. doi:10.3389/fmicb.2014.00141. PMC 3983520. PMID 24772109.
- Gómez-Gómez L, Boller T (June 2000). "FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis". Molecular Cell. 5 (6): 1003–11. doi:10.1016/S1097-2765(00)80265-8. PMID 10911994.
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T (July 2007). "A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence". Nature. 448 (7152): 497–500. Bibcode:2007Natur.448..497C. doi:10.1038/nature05999. hdl:11858/00-001M-0000-0012-3840-F. PMID 17625569.
External links
- Flagellin at the US National Library of Medicine Medical Subject Headings (MeSH)
- Bacterial flagellin and plant disease resistance, published by Zipfel. et al. (2004) Abstract Article