Archaerhodopsin
Archaerhodopsin (alternatively known as bacterio-opsin) is a family of receptor proteins that respond to light, found in Archaea. They function similarly to the mammalian protein Rhodopsin, but are structurally distinct. Archaerhodopsins are widely used as optogenetics tools for neuroscience research.
Gene
Archaerhodopsins are light-driven outward proton pump retinal proteins, similar to bacteriorhodopsins,.[1] Archaerododhopsin 1 and 2 (aR-1 and aR-2) represent the first archaerhodopsins founded in Halobacterium family isolated from Australian strains by Mukahata et al., 1988, and Uegaki et al., 1991. Archaerododhopsin 1 and 2 (aR-1 and aR-2), are present in Halobacterial species, Halobacterium sp. Aus-1[2] and Aus-2[3] respectively.
In addition to (aR-1) and (aR-2), Archaerhodopsin-3 (aR-3), is another microbial rhodopsin proton pump founded in Halorubrum sodomense species. That has recently been introduced as a fluorescent voltage sensor.[4] The fourth Archaerhodopsin, Archaerodopsin-4 (aR-4), represents a new member of the microbial rhodopsin family founded in Halobacterium species xz 515 collected from a salt take in Tibet,.[5][6]
Archaerhodopsin proteins mentioned above carried the some ion pumps function but with different amino acid sequences which reflects difference in Halobacteria strains and related ecological habitats. For example, according to Sugiyama et al., 1989 and Ubgaki et al., 1991, the amino acid sequences which were deduced from the structural genes coding the polypeptides of both (aR-1) an (aR-2) indicated that all essential residues are conserved,.[7][3] While Archaerhodopsin-3 protein encoded by aop3 gene is closely related to Archaerodopsin-1 (aR-1) and Archaerodopsin-2 (aR-2) with over 90% homology.[8] However, (aR-4), was found membrane of Halobacterium species xz 515 collected from a salt take in Tibet, shows 87% sequence similarity to aR-1, 97% to aR-2 and 84% to aR-3. Archaerhodopsin gene of xz 515 encoding the protein from helix C to helix G has been identified by H. Wang et al., 2000.[9] The genes encoding archaerhodopsin proteins and its related Halobacterium strains, origins and subcelular location are illustrated in the present table:
- Archaerhodopsin-1 (AR-1)
- Archaerhodopsin-2 (AR-2)
- Archaerhodopsin-3 (AR-3)
- Archaerhodopsin-BD1 (AR-BD1)
- Archaerhodopsin-4 (AR-4)
Structure
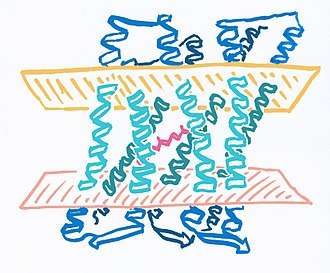
Archaerhodopsin a is a protein family composed of 258 to 260 amino acids, depending on the type (AR-1, AR-2, AR-3, AR-4, AR-BD1). These form extracellular, cytoplasmic and helix alpha transmembrane domains, as are multi-pass membrane proteins.[10]
As previously mentioned archaerhodopsin proteic family differentiates itself from the bacteriorhodopsin (bR) proteins by a 55-58% of its primary structure on average.[11]
Differences among archaerodopsine reside also in the location of these. As for example AR-1 is found in Halobacterium sp. SG1 and Halorubrum chaoviator, while AR-2 in Halobacterium sp. AUS-2, AR-3 in Halorubrum sodomense, and AR-BD1 in Halorubrum xinjiangense.
Diagram Key
- Light/Mint blue: Transmembrane domain
- Dark blue: Cytoplasmic and extracellular domains.
- Magenta: Opsin-bound retinal.
- Light pink: Cytoplasmatic region of cell membrane.
- Orange: Extracelular region of cell membrane.
Function
Archaerhodopsins act primarily as light-driven proton pumps. They transduce a light signal to change the protein shape and allow the release of protons by using the pigment retinal, similarly to other Opsin proteins. Thus it has two distinct activities: photoreceptor activity and ion channel activity. The biological processes that undertakes thus are the transduction of light stimuli, the transport of protons and the protein-chromophore linkage.[10]
The retinal proteins, bacteriorhodopsin (bR)’ and halorhodopsin (hR) of Halobacterium halobium, are light-driven pumps for the transport of H’ and C1- ions respectively.
Use in research
Archaerhodopsins drive the hyperpolarization of the cell membrane by secreting protons in presence of light, thereby inhibiting action potential firing of neurons.[12] This process is associated to an increase in cytosolic pH linked to the activity of these proteins. These characteristics allow for Archaerhodopsins to be commonly used tools for optogenetic studies as they behave as transmission inhibition factors in presence of light.[13] The proton pump activity can also be used for optogenetic acidification of lysosomes and synaptic vesicles when targeted to these organelles.[14]
History
In the 1960s, a light driven proton pump was discovered in Halobacterium salinarum, and called Bacteriorhodopsin. Over the following years, there were various studies of the membrane of H.salinarum to determine the mechanism of these light-driven proton pumps.
In 1988, another Manabu Yoshida's group at Osaka University reported a novel light-sensitive proton pump from a strain of Halobacterium which they termed Archaerhodopsin.[2] A year later, the same group reported isolating the gene that encodes Archaerhodopsin.[15][16]
Etymology
Archaerhodopsin is named after Archaea, the domain in which the protein family was first found, and Rhodopsin (commonly known as visual purple), the protein family Archaerhodopsins belong to.
Literature
- Inoue K, Ito S, Kato Y, Nomura Y, Shibata M, Uchihashi T, Tsunoda SP, Kandori H (November 2016). "A natural light-driven inward proton pump". Nature Communications. 7: 13415. doi:10.1038/ncomms13415. PMC 5118547. PMID 27853152.
- Mukohata Y, Sugiyama Y, Ihara K, Yoshida M (March 1988). "An Australian halobacterium contains a novel proton pump retinal protein: archaerhodopsin". Biochemical and Biophysical Research Communications. 151 (3): 1339–45. doi:10.1016/S0006-291X(88)80509-6. PMID 2833260.
- Uegaki K, Sugiyama Y, Mukohata Y (April 1991). "Archaerhodopsin-2, from Halobacterium sp. aus-2 further reveals essential amino acid residues for light-driven proton pumps". Archives of Biochemistry and Biophysics. 286 (1): 107–10. doi:10.1016/0003-9861(91)90014-A. PMID 1654776.
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE (November 2011). "Optical recording of action potentials in mammalian neurons using a microbial rhodopsin". Nature Methods. 9 (1): 90–5. doi:10.1038/nmeth.1782. PMC 3248630. PMID 22120467.
- Li Q, Sun Q, Zhao W, Wang H, Xu D (June 2000). "Newly isolated archaerhodopsin from a strain of Chinese halobacteria and its proton pumping behavior". Biochimica et Biophysica Acta. 1466 (1–2): 260–6. doi:10.1016/S0005-2736(00)00188-7. PMID 10825447.
- Y. Wang, D. Ma, Y. Zhao, M. Ming, J. Wu, J., 2012. Ding Light-driven proton pumps of archaerhodopsin and bacteriorhodopsin & polymer-matrix composite materials of those functional proteins. Acta Polym. Sin. 698-713.
- Sugiyama Y, Maeda M, Futai M, Mukohata Y (December 1989). "Isolation of a gene that encodes a new retinal protein, archaerhodopsin, from Halobacterium sp. aus-1". The Journal of Biological Chemistry. 264 (35): 20859–62. PMID 2592356.
- Ihara K, Umemura T, Katagiri I, Kitajima-Ihara T, Sugiyama Y, Kimura Y, Mukohata Y (January 1999). "Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation". Journal of Molecular Biology. 285 (1): 163–74. doi:10.1006/jmbi.1998.2286. PMID 9878396.
- . Wang, S. Zhan, Q. Sun, D. Xu, W. Zhao, W. Huang, Q. Li., 2000. Primary structure of helix C to helix G of a new retinal protein in H.sp.xz515. Chin. Sci. Bull., 45: 1108-1113.
- Archaerhodopsin in UniProtKB UniProt Knowledgebase
- Enami N, Yoshimura K, Murakami M, Okumura H, Ihara K, Kouyama T (May 2006). "Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps". Journal of Molecular Biology. 358 (3): 675–85. doi:10.1016/j.jmb.2006.02.032. PMID 16540121.
- Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES (January 2010). "High-performance genetically targetable optical neural silencing by light-driven proton pumps". Nature. 463 (7277): 98–102. doi:10.1038/nature08652. PMC 2939492. PMID 20054397.
- Mohamady El-Gaby, Yu Zhang, Konstantin Wolf, Christof J. Schwiening, Ole Paulsen, Olivia A. Shipton. 2016. Archaerhodopsin Selectively and Reversibly Silences Synaptic Transmission through Altered pH. Cell Reports 16(8):2259-2268
- Rost BR, Schneider F, Grauel MK, Wozny C, Bentz C, Blessing A, Rosenmund T, Jentsch TJ, Schmitz D, Hegemann P, Rosenmund C (December 2015). "Optogenetic acidification of synaptic vesicles and lysosomes". Nature Neuroscience. 18 (12): 1845–1852. doi:10.1038/nn.4161. PMC 4869830. PMID 26551543.
- Sugiyama Y, Maeda M, Futai M, Mukohata Y (December 1989). "Isolation of a gene that encodes a new retinal protein, archaerhodopsin, from Halobacterium sp. aus-1". The Journal of Biological Chemistry. 264 (35): 20859–62. PMID 2592356.
- Oren, A (1994). "Enzyme diversity in halophilic archaea" (PDF). Microbiología. 10: 217–228. Retrieved 8 April 2018.
- Archaea in Wiktionary The free dictionary
- Rhodopsin in Wiktionary The free dictionary