Venom
Venom is a secretion containing one or more toxins produced by an animal.[1] Venom has evolved in a wide variety of animals, both predators and prey, and both vertebrates and invertebrates.

Venoms kill through the action of at least four major classes of toxin, namely necrotoxins and cytotoxins, which kill cells; neurotoxins, which affect nervous systems; and myotoxins, which damage muscles. Biologically, venom is distinguished from poison in that poisons are ingested, while venom is delivered in a bite, sting, or similar action. Venomous animals cause tens of thousands of human deaths per year. However, the toxins in many venoms have potential to treat a wide range of diseases.
Evolution
The use of venom across a wide variety of taxa is an example of convergent evolution. It is difficult to conclude exactly how this trait came to be so intensely widespread and diversified. The multigene families that encode the toxins of venomous animals are actively selected, creating more diverse toxins with specific functions. Venoms adapt to their environment and victims and accordingly evolve to become maximally efficient on a predator's particular prey (particularly the precise ion channels within the prey). Consequently, venoms become specialized to an animal's standard diet.[2]
Mechanisms
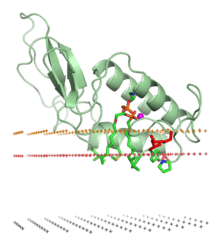
Venoms cause their biological effects via the toxins that they contain; some venoms are complex mixtures of toxins of differing types. Among the major classes of toxin in venoms are:[3]
- Necrotoxins, which cause necrosis (i.e., death) in the cells they encounter.[4] The venom of most viper species contains phospholipase and trypsin-like serine proteases.
- Neurotoxins, which primarily affect the nervous systems of animals.[5] These include ion channel toxins that disrupt ion channel conductance. Black widow spider, scorpion, box jellyfish, cone snail, centipede and blue-ringed octopus venoms (among many others) function in this way.
- Myotoxins, which damage muscles by binding to a receptor, are small, basic peptides found in snake (such as rattlesnake) and lizard venoms.[6][7][8][9]
- Cytotoxins, which kill individual cells, are found in the apitoxin of honey bees and the venom of black widow spiders.[10][11]
Taxonomic range

Venom is widely distributed taxonomically, being found in both invertebrates and vertebrates; in aquatic and terrestrial animals; and among both predators and prey. The major groups of venomous animals are described below.
Arthropods
Venomous arthropods include spiders, which use fangs — part of their chelicerae — to inject venom; and centipedes, which use forcipules — modified legs — to deliver venom; along with scorpions and stinging insects, which inject venom with a sting.
In insects such as bees and wasps, the stinger is a modified egg-laying device — the ovipositor. In Polistes fuscatus, the female continuously releases a venom that contains a sex pheromone that induces copulatory behavior in males.[12] In Polistes exclamans, venom is used as an alarm pheromone, coordinating a response with from the nest and attracting nearby wasps to attack the predator.[13] In Dolichovespula arenaria, the observed spraying of venom out of their sting has been seen from workers in large colonies.[14] In other cases like Parischnogaster striatula, the venom is applied all over their body as an antimicrobial protection.[15] The venom from Agelaia pallipes has inhibitory effects on processes like chemotaxis and hemolysis which can lead to organ failure.[16]
Many caterpillars have defensive venom glands associated with specialized bristles on the body, known as urticating hairs, which can be lethal to humans (e.g., that of the Lonomia moth), although the venom's strength varies depending on the species.[17]
Bees synthesize and employ an acidic venom (apitoxin) to cause pain in those that they sting to defend their hives and food stores, whereas wasps use a chemically different alkaline venom designed to paralyze prey, so it can be stored alive in the food chambers of their young. The use of venom is much more widespread than just these examples. Other insects, such as true bugs and many ants, also produce venom.[18] At least one ant species (Polyrhachis dives) has been shown to use venom topically for the sterilisation of pathogens.[19]
Other invertebrates
There are venomous invertebrates in several phyla, including jellyfish such as the dangerous box jellyfish[20] and sea anemones among the Cnidaria,[21] sea urchins among the Echinodermata,[22] and cone snails[23] and cephalopods including octopuses among the Molluscs.[24]
Vertebrates
Fish
Venom is found in some 200 cartilaginous fishes, including stingrays, sharks, and chimaeras; the catfishes (about 1000 venomous species); and 11 clades of spiny-rayed fishes (Acanthomorpha), containing the scorpionfishes (over 300 species), stonefishes (over 80 species), gurnard perches, blennies, rabbitfishes, surgeonfishes, some velvetfishes, some toadfishes, coral crouchers, red velvetfishes, scats, rockfishes, deepwater scorpionfishes, waspfishes, weevers, and stargazers.[25]
Reptiles

Some 450 species of snake are venomous.[25] Snake venom is produced by glands below the eye (the mandibular gland) and delivered to the victim through tubular or channeled fangs. Snake venoms contain a variety of peptide toxins, including proteases, which hydrolyze protein peptide bonds, nucleases, which hydrolyze the phosphodiester bonds of DNA, and neurotoxins, which disable signalling in the nervous system.[28] Snake venom causes symptoms including pain, swelling, tissue necrosis, low blood pressure, convulsions, hemorrhage (varying by species of snake), respiratory paralysis, kidney failure, coma and death.[29] Snake venom may have originated with duplication of genes that had been expressed in the salivary glands of ancestors.[30][31]
Venom is found in a few other reptiles such as the Mexican beaded lizard,[32] the gila monster,[33] and some monitor lizards including the Komodo dragon.[34] Mass spectrometry showed that the mixture of proteins present in their venom is as complex as the mixture of proteins found in snake venom.[34][35] Some lizards possess a venom gland; they form a hypothetical clade, Toxicofera, containing the suborders Serpentes and Iguania and the families Varanidae, Anguidae, and Helodermatidae.[36]
Mammals
Euchambersia, an extinct genus of therocephalians, is hypothesized to have had venom glands attached to its canine teeth.[37]
A few species of living mammals are venomous, including solenodons, shrews, vampire bats, the male platypus and the slow loris.[25][38] Shrews are known to have venomous saliva and most likely evolved their trait similarly to snakes.[39] The presence of tarsal spurs akin to those of the platypus in many non-therian Mammaliaformes groups suggests that venom was an ancestral characteristic among mammals.[40]
Extensive research on platypuses shows that their toxin was initially formed from gene duplication, but data provides evidence that the further evolution of platypus venom does not rely as much on gene duplication as once was thought.[41] Modified sweat glands are what evolved into platypus venom glands. Although it is proven that reptile and platypus venom have independently evolved, it is thought that there are certain protein structures that are favored to evolve into toxic molecules. This provides more evidence as to why venom has become a homoplastic trait and why very different animals have convergently evolved.[42]
Venom and humans
Venomous animals resulted in 57,000 human deaths in 2013, down from 76,000 deaths in 1990.[43]
Venoms, found in over 173,000 species, have potential to treat a wide range of diseases, explored in over 5,000 scientific papers.[33] Snake venoms contain proteins which can be used to treat conditions including thrombosis, arthritis, and some cancers.[44][45] Gila monster venom contains exenatide, used to treat type 2 diabetes.[33]
Solenopsins extracted from fire ant venom has demonstrated biomedical applications, ranging from cancer treatment to psoriasis.[46][47]
Venom Resistance
Coevolutionary Adaptations
Venom is utilized as a trophic weapon by multiple predator species. The coevolution between predators and prey is a driving force of venom resistance, which has evolved multiple times throughout the animal kingdom. Repeated interactions between two species can generate coevolution.[48] The coevolution between venomous predators and venom resistant prey is best described as a chemical arms race.[49] Predator and prey pairs are expected to associate with one another for stable periods of time.[50] Venom is used as a chemical weapon by predator species. As the predator capitalizes on susceptible individuals, the surviving individuals are limited to those who are able to evade predation.[51] Resistance phenotypes typically increase over time as the predator becomes increasingly unable to subdue prey that have developed this new resistance phenotype.[52]
The cost of developing a venom resistance is high, for predator and prey.[53] Developing an entire physiological resistance is extremely costly, however it maximizes chances of survival for prey species and allows predator species to expand into underutilized trophic niches. If it is possible for an animal to evade predation through something less costly like a behavioral modification, the development of a physiological modification becomes unnecessary.[54]
Venom Resistance in Vertebrates
California Ground Squirrel and Northern Pacific Rattlesnake
_(6-19-10)_cuesta_ridge%2C_slo_co%2C_ca_-04_(4715157755).jpg)
One of the most heavily researched cases of venom resistance is the California Ground Squirrel, which is resistant to the venom of the Northern Pacific Rattlesnake. The predator-prey pair have coexisted for generations. The repeated interactions fostered the development of an anti-snake venom defense in the California ground squirrels. Researchers found evidence supporting the theory that venom resistance is driven by coevolution among these populations of California ground squirrels.[55] They use toxin scavenging to negate the effects of the haemolytic toxins of their rattlesnake predators, demonstrating a physiological resistance to rattlesnake venom. Resistance in these ground squirrels is population dependent. In areas where rattlesnake populations are very dense, there is a significant increase in resistance in the squirrels compared to populations where rattlesnakes are rare.[56] Rattlesnakes demonstrated local adaptions in the effectiveness of their venom in order to overcome the venom resistant squirrels.[57]
Eels and Sea Snakes
There is an ongoing evolutionary contest between toxicity and resistance. The resistance of eels to sea snake venom is a good example of coevolution between predator-prey pairs. Sea snake venom is composed of complex mixtures of neurotoxins, myotoxins, nephrotoxins, and other nontoxic substances.[58] The composition of the sea snake venom is species specific. The biggest piece of evidence for this as a case of coevolution is that eels that are favored by sea snakes as prey have unusually high tolerances to the venom of the sea snake.[59] Studies have analyzed the resistance of four species of eels to two different sea snakes: one is a dietary generalist and the other is an eel specialist.[60] The eels were more resistant to the venom of the eel specialist sea snake. Non-prey fishes exhibited very low levels of resistance to the sea snake venom, further supporting coevolution.[60]
Clownfish and Sea Anemones
Clownfish make their homes in the safety among the tentacles of sea anemones. Sea anemones are one of the most venomous organisms on the planet, yet clownfish are able to inhabit the sea anemone unharmed.[61] The genetic mechanisms allowing clownfish to interact with sea anemones is still somewhat unclear.[62] Only 10 known species of anemones are hosts to clownfish and only certain pairs of anemones and clownfish are compatible with one another.[63][64]

All sea anemones produce venoms delivered through discharging nematocysts and mucous secretions. The toxins are composed of peptides and proteins. They are used for prey acquisition and to deter predators by causing pain, loss of muscular coordination and tissue damage. Clownfish have a protective mucous that acts as a chemical camouflage or macromolecular mimicry preventing “not self” recognition by the sea anemone and nematocyst discharge.[65] The sea anemone perceives the fish as its “self” most likely by the same mechanism which prevents the discharge of nematocysts when its tentacles come into contact with each other.[66] Clownfish exhibit either strict host specificity or are environmental niche generalists and can associate with a variety of sea anemone species.[67] In some species, the mucus of clownfish was found to change during acclimation to resemble that of the specific species of sea anemone.[67] For the Clownfish, a relationship with the sea anemone is an obligatory one. In some cases, it is an obligatory relationship for the anemone as well. In all cases, the interaction between the two is mutually beneficial. Clownfish and sea anemones are one of the most compelling cases of symbiosis. The relationship provides mutual protection from predators and the exchange of nutrients. Divergent natural selection drives adaptive diversification through ecological speciation. The ability of clownfish to use sea anemones as hosts has evolved 4 independent times. The obligate relationship between the clownfish and the sea anemone has allowed the radiation of clownfishes.[68]
Kingsnakes
Inhabiting the Americas from southeastern Canada to southern Ecuador, Kingsnakes, of genus Lampropeltis, are constrictors that prey on many venomous snakes.[69] In order to prey upon the venomous snakes, Kingsnakes have actually evolved a resistance rather than incrementally increasing their resistance to a point of immunity, like many species have. Kingsnakes resistance levels are currently known to be fixed for the duration of its life and have not been found to change with age or exposure. It is thought, that Kingsnakes have developed this evolutionary adaptation through a process called a co-evolutionary arms race with natural selection at the forefront.[70] Kingsnake predators that were slightly better able to tolerate the effects of the venom were more likely to survive with resulting genocide of the Kingsnakes that were inherently not sufficiently resistant. While simultaneously, the venomous snakes with more potent venom were more likely to survive the predatory nature of the Kingsnakes, thus escalating the arms race.
However, the nature of the arms race has put a stipulation onto the Kingsnakes. Kingsnakes have evolved resistance only to the venom of snakes that are in their immediate environment, like copperheads, cottonmouths, and North American rattlesnakes, but not to the venom of, for example, king cobras or black mambas. Even through geographical boundaries, Kingsnake venom resistance has varied between species. They found that blood from Eastern Kingsnakes (Lampropeltis getula) had the widest spectrum of protection against the venoms tested and was the most effective at neutralizing many rattlesnake venoms, but the least effective against copperhead venom. Blood from kingsnakes from Florida & the Gulf Coast was the most effective at neutralizing the venom of copperheads & cottonmouths. Mole Kingsnake (Lampropeltis calligaster) blood is about 75% as effective at neutralizing Mojave Rattlesnake (Crotalus scutulatus) venom as the blood of Eastern Kingsnakes. Gray-banded Kingsnakes (L. alterna) have moderate neutralization potential against Western Diamondback (C. atrox) venom, but none against Eastern Diamondback (C. adamanteus) venom.[71]
See also
- Poison
- Envenomation
- Schmidt Sting Pain Index
- Big Four (Indian snakes)
- List of venomous animals
- Venomous mammals
- Venoms in medicine
References
- "venom" at Dorland's Medical Dictionary
- Kordiš, D.; Gubenšek, F. (2000). "Adaptive evolution of animal toxin multigene families". Gene. 261 (1): 43–52. doi:10.1016/s0378-1119(00)00490-x. PMID 11164036.
- Harris, J. B. (September 2004). "Animal poisons and the nervous system: what the neurologist needs to know". Journal of Neurology, Neurosurgery & Psychiatry. 75 (suppl_3): iii40–iii46. doi:10.1136/jnnp.2004.045724. PMC 1765666. PMID 15316044.
- Raffray M, Cohen GM; Cohen (1997). "Apoptosis and necrosis in toxicology: a continuum or distinct modes of cell death?". Pharmacol. Ther. 75 (3): 153–177. doi:10.1016/s0163-7258(97)00037-5. PMID 9504137.
- Dutertre, Sébastien; Lewis, Richard J. (2006). "Toxin insights into nicotinic acetylcholine receptors". Biochemical Pharmacology. 72 (6): 661–670. doi:10.1016/j.bcp.2006.03.027. PMID 16716265.
- Nicastro, G; Franzoni, L; de Chiara, C; Mancin, A. C.; Giglio, J. R.; Spisni, A. (May 2003). "Solution structure of crotamine, a Na+ channel affecting toxin from Crotalus durissus terrificus venom". Eur. J. Biochem. 270 (9): 1969–1979. doi:10.1046/j.1432-1033.2003.03563.x. PMID 12709056.CS1 maint: multiple names: authors list (link)
- Griffin, P. R.; Aird, S. D. (1990). "A new small myotoxin from the venom of the prairie rattlesnake (Crotalus viridis viridis)". FEBS Lett. 274 (1): 43–47. doi:10.1016/0014-5793(90)81325-I. PMID 2253781.CS1 maint: multiple names: authors list (link)
- Samejima Y.; Aoki, Y; Mebs, D. (1991). "Amino acid sequence of a myotoxin from venom of the eastern diamondback rattlesnake (Crotalus adamanteus)". Toxicon. 29 (4): 461–468. doi:10.1016/0041-0101(91)90020-r. PMID 1862521.CS1 maint: multiple names: authors list (link)
- Whittington, C. M.; Papenfuss, A. T.; Bansal, P.; Torres, A. M.; Wong, E. S.; Deakin, J. E.; Graves, T.; Alsop, A.; Schatzkamer, K.; Kremitzki, C.; Ponting, C. P.; Temple-Smith, P.; Warren, W. C.; Kuchel, P. W.; Belov, K. (June 2008). "Defensins and the convergent evolution of platypus and reptile venom genes". Genome Research. 18 (6): 986–094. doi:10.1101/gr.7149808. PMC 2413166. PMID 18463304.
- Sobral, Filipa; Sampaio, Andreia; Falcão, Soraia; Queiroz, Maria João R.P.; Calhelha, Ricardo C.; Vilas-Boas, Miguel; Ferreira, Isabel C.F.R. (2016). "Chemical characterization, antioxidant, anti-inflammatory and cytotoxic properties of bee venom collected in Northeast Portugal" (PDF). Food and Chemical Toxicology. 94: 172–177. doi:10.1016/j.fct.2016.06.008. hdl:10198/13492. PMID 27288930.
- Peng, Xiaozhen; Dai, Zhipan; Lei, Qian; Liang, Long; Yan, Shuai; Wang, Xianchun (April 2017). "Cytotoxic and apoptotic activities of black widow spiderling extract against HeLa cells". Experimental and Therapeutic Medicine. 13 (6): 3267–3274. doi:10.3892/etm.2017.4391. PMC 5450530. PMID 28587399.
- Post David, Jeanne Robert (1983). "Venom: Source of a Sex Pheromone in the Social Wasp Polistes fuscatus (Hymenoptera: Vespidae)". Journal of Chemical Ecology. 9 (2): 259–266. doi:10.1007/bf00988043. PMID 24407344.
- Post Downing, Jeanne (1984). "Alarm response to venom by social wasps Polistes exclamans and P. fuscatus". Journal of Chemical Ecology. 10 (10): 1425–1433. doi:10.1007/BF00990313. PMID 24318343.
- Greene, Alex. "The Aerial Yellowjacket Dolichovespula Arenaria." Academia.edu. Department of Entomology — Washington State University, n.d. Web. 25 September 2014.
- Baracchi, David (January 2012). "From individual to collective immunity: The role of the venom as antimicrobial agent in the Stenogastrinae wasp societies". Journal of Insect Physiology. 58 (1): 188–193. doi:10.1016/j.jinsphys.2011.11.007. hdl:2158/790328. PMID 22108024.
- Baptista-Saidemberg, Nicoli; et al. (2011). "Profiling the peptidome of the venom from the social wasp Agelaia pallipes pallipes". Journal of Proteomics. 74 (10): 2123–2137. doi:10.1016/j.jprot.2011.06.004. PMID 21693203.
- Pinto, Antônio F. M.; Berger, Markus; Reck, José; Terra, Renata M. S.; Guimarães, Jorge A. (15 December 2010). "Lonomia obliqua venom: In vivo effects and molecular aspects associated with the hemorrhagic syndrome". Toxicon. 56 (7): 1103–1112. doi:10.1016/j.toxicon.2010.01.013. PMID 20114060.
- Touchard, Axel; Aili, Samira; Fox, Eduardo; Escoubas, Pierre; Orivel, Jérôme; Nicholson, Graham; Dejean, Alain (20 January 2016). "The Biochemical Toxin Arsenal from Ant Venoms". Toxins. 8 (1): 30. doi:10.3390/toxins8010030. ISSN 2072-6651. PMC 4728552. PMID 26805882.
- Graystock, Peter; Hughes, William O. H. (2011). "Disease resistance in a weaver ant, Polyrhachis dives, and the role of antibiotic-producing glands". Behavioral Ecology and Sociobiology. 65 (12): 2319–2327. doi:10.1007/s00265-011-1242-y.
- Frost, Emily (30 August 2013). "What's Behind That Jellyfish Sting?". Smithsonian. Retrieved 30 September 2018.
- Bonamonte, Domenico; Angelini, Gianni (2016). Aquatic Dermatology: Biotic, Chemical and Physical Agents. Springer International. pp. 54–56. ISBN 978-3-319-40615-2.
- Gallagher, Scott A. (2 August 2017). "Echinoderm Envenomation". EMedicine. Retrieved 12 October 2010.
- Olivera, B. M.; Teichert, R. W. (2007). "Diversity of the neurotoxic Conus peptides: a model for concerted pharmacological discovery". Molecular Interventions. 7 (5): 251–260. doi:10.1124/mi.7.5.7. PMID 17932414.
- Barry, Carolyn (17 April 2009). "All Octopuses Are Venomous, Study Says". National Geographic. Retrieved 30 September 2018.
- Smith, William Leo; Wheeler, Ward C. (2006). "Venom Evolution Widespread in Fishes: A Phylogenetic Road Map for the Bioprospecting of Piscine Venoms". Journal of Heredity. 97 (3): 206–217. doi:10.1093/jhered/esj034. PMID 16740627.
- Venomous Amphibians (Page 1) - Reptiles (Including Dinosaurs) and Amphibians - Ask a Biologist Q&A. Askabiologist.org.uk. Retrieved on 2013-07-17.
- Nowak, R. T.; Brodie, E. D. (1978). "Rib Penetration and Associated Antipredator Adaptations in the Salamander Pleurodeles waltl (Salamandridae)". Copeia. 1978 (3): 424–429. doi:10.2307/1443606. JSTOR 1443606.
- Bauchot, Roland (1994). Snakes: A Natural History. Sterling. pp. 194–209. ISBN 978-1-4027-3181-5.
- "Snake Bites". A. D. A. M. Inc. 16 October 2017. Retrieved 30 September 2018.
- Hargreaves, Adam D.; Swain, Martin T.; Hegarty, Matthew J.; Logan, Darren W.; Mulley, John F. (30 July 2014). "Restriction and Recruitment—Gene Duplication and the Origin and Evolution of Snake Venom Toxins". Genome Biology and Evolution. 6 (8): 2088–2095. doi:10.1093/gbe/evu166. PMC 4231632. PMID 25079342.
- Daltry, Jennifer C.; Wuester, Wolfgang; Thorpe, Roger S. (1996). "Diet and snake venom evolution". Nature. 379 (6565): 537–540. doi:10.1038/379537a0. PMID 8596631.
- Cantrell, F. L. (2003). "Envenomation by the Mexican beaded lizard: a case report". Journal of Toxicology. Clinical Toxicology. 41 (3): 241–244. doi:10.1081/CLT-120021105. PMID 12807305.
- Mullin, Emily (29 November 2015). "Animal Venom Database Could Be Boon To Drug Development". Forbes. Retrieved 30 September 2018.
- Fry, B. G.; Wroe, S.; Teeuwisse, W.; et al. (June 2009). "A central role for venom in predation by Varanus komodoensis (Komodo Dragon) and the extinct giant Varanus (Megalania) priscus". PNAS. 106 (22): 8969–8974. doi:10.1073/pnas.0810883106. PMC 2690028. PMID 19451641.
- Fry, B. G.; Wuster, W.; Ramjan, S. F. R.; Jackson, T.; Martelli, P.; Kini, R. M. 2003c. Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Communications in Mass Spectrometry 17:2047-2062.
- Fry, B. G.; Vidal, N.; Norman, J. A.; Vonk, F. J.; Scheib, H.; Ramjan, S. F.; Kuruppu, S.; Fung, K.; Hedges, S. B.; Richardson, M. K.; Hodgson, W. C.; Ignjatovic, V.; Summerhayes, R.; Kochva, E. (February 2006). "Early evolution of the venom system in lizards and snakes". Nature. 439 (7076): 584–588. doi:10.1038/nature04328. PMID 16292255.
- Benoit, J.; Norton, L. A.; Manger, P. R.; Rubidge, B. S. (2017). "Reappraisal of the envenoming capacity of Euchambersia mirabilis (Therapsida, Therocephalia) using μCT-scanning techniques". PLoS ONE. 12 (2): e0172047. doi:10.1371/journal.pone.0172047. PMC 5302418. PMID 28187210.
- Nekaris, K. Anne-Isola; Moore, Richard S.; Rode, E. Johanna; Fry, Bryan G. (27 September 2013). "Mad, bad and dangerous to know: the biochemistry, ecology and evolution of slow loris venom". Journal of Venomous Animals and Toxins Including Tropical Diseases. 19 (1): 21. doi:10.1186/1678-9199-19-21. PMC 3852360. PMID 24074353.
- Ligabue-Braun, R.; Verli, H.; Carlini, C. R. (2012). "Venomous mammals: a review". Toxicon. 59 (7–8): 680–695. doi:10.1016/j.toxicon.2012.02.012. PMID 22410495.
- Jørn H. Hurum, Zhe-Xi Luo, and Zofia Kielan-Jaworowska, Were mammals originally venomous?, Acta Palaeontologica Polonica 51 (1), 2006: 1-11
- Wong, E. S.; Belov, K. (2012). "Venom evolution through gene duplications". Gene. 496 (1): 1–7. doi:10.1016/j.gene.2012.01.009. PMID 22285376.
- Whittington C. M.; Papenfuss A. T.; Bansal P.; Torres A. M.; Wong E. S.; Deakin J. E.; Belov K. (2008). "Defensins and the convergent evolution of platypus and reptile venom genes". Genome Research. 18 (6): 986–994. doi:10.1101/gr.7149808. PMC 2413166. PMID 18463304.
- GBD 2013 Mortality and Causes of Death, Collaborators (17 December 2014). "Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013". Lancet. 385 (9963): 117–171. doi:10.1016/S0140-6736(14)61682-2. PMC 4340604. PMID 25530442.
- Pal, S. K.; Gomes, A.; Dasgupta, S. C.; Gomes, A. (2002). "Snake venom as therapeutic agents: from toxin to drug development". Indian Journal of Experimental Biology. 40 (12): 1353–1358. PMID 12974396.
- Holland, Jennifer S. (February 2013). "The Bite That Heals". National Geographic. Retrieved 30 September 2018.
- Fox, Eduardo G.P.; Xu, Meng; Wang, Lei; Chen, Li; Lu, Yong-Yue (May 2018). "Speedy milking of fresh venom from aculeate hymenopterans". Toxicon. 146: 120–123. doi:10.1016/j.toxicon.2018.02.050. PMID 29510162.
- Fox, Eduardo Gonçalves Paterson (2021), Gopalakrishnakone, P.; Calvete, Juan J. (eds.), "Venom Toxins of Fire Ants", Venom Genomics and Proteomics: Venom Genomics and Proteomics, Springer Netherlands, pp. 1–16, doi:10.1007/978-94-007-6649-5_38-1 (inactive 2019-08-20), ISBN 9789400766495 Check date values in:
|date=(help) - Arbuckle, Kevin; Rodríguez de la Vega, Ricardo C.; Casewell, Nicholas R. (December 2017). "Coevolution takes the sting out of it: Evolutionary biology and mechanisms of toxin resistance in animals". Toxicon. 140: 118–131. doi:10.1016/j.toxicon.2017.10.026.
- Dawkins, Richard; Krebs, John Richard; Maynard Smith, J.; Holliday, Robin (21 September 1979). "Arms races between and within species". Proceedings of the Royal Society of London. Series B. Biological Sciences. 205 (1161): 489–511. doi:10.1098/rspb.1979.0081.
- McCabe, Thomas M.; Mackessy, Stephen P. (2015), Gopalakrishnakone, P.; Malhotra, Anita (eds.), "Evolution of Resistance to Toxins in Prey", Evolution of Venomous Animals and Their Toxins, Toxinology, Springer Netherlands, pp. 1–19, doi:10.1007/978-94-007-6727-0_6-1, ISBN 978-94-007-6727-0, retrieved 1 December 2019
- Nuismer, Scott L.; Ridenhour, Benjamin J.; Oswald, Benjamin P. (2007). "Antagonistic Coevolution Mediated by Phenotypic Differences Between Quantitative Traits". Evolution. 61 (8): 1823–1834. doi:10.1111/j.1558-5646.2007.00158.x. ISSN 1558-5646.
- Holding, Matthew L.; Drabeck, Danielle H.; Jansa, Sharon A.; Gibbs, H. Lisle (1 November 2016). "Venom Resistance as a Model for Understanding the Molecular Basis of Complex Coevolutionary Adaptations". Integrative and Comparative Biology. 56 (5): 1032–1043. doi:10.1093/icb/icw082. ISSN 1540-7063.
- Calvete, Juan J. (1 March 2017). "Venomics: integrative venom proteomics and beyond". Biochemical Journal. 474 (5): 611–634. doi:10.1042/BCJ20160577. ISSN 0264-6021.
- Morgenstern, David; King, Glenn F. (1 March 2013). "The venom optimization hypothesis revisited". Toxicon. 63: 120–128. doi:10.1016/j.toxicon.2012.11.022. ISSN 0041-0101.
- Poran, Naomie S.; Coss, Richard G.; Benjamini, Eli (1 January 1987). "Resistance of California ground squirrels (Spermophilus Beecheyi) to the venom of the northern Pacific rattlesnake (Crotalus Viridis Oreganus): A study of adaptive variation". Toxicon. 25 (7): 767–777. doi:10.1016/0041-0101(87)90127-9. ISSN 0041-0101.
- Coss, Richard G.; Poran, Naomie S.; Gusé, Kevin L.; Smith, David G. (1 January 1993). "Development of Antisnake Defenses in California Ground Squirrels (Spermophilus Beecheyi): II. Microevolutionary Effects of Relaxed Selection From Rattlesnakes". Behaviour. 124 (1–2): 137–162. doi:10.1163/156853993X00542. ISSN 0005-7959.
- Holding, Matthew L.; Biardi, James E.; Gibbs, H. Lisle (27 April 2016). "Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey". Proceedings of the Royal Society B: Biological Sciences. 283 (1829): 20152841. doi:10.1098/rspb.2015.2841. PMC 4855376. PMID 27122552.
- Heatwole, Harold; Poran, Naomie S. (15 February 1995). "Resistances of Sympatric and Allopatric Eels to Sea Snake Venoms". Copeia. 1995 (1): 136. doi:10.2307/1446808.
- Heatwole, Harold; Powell, Judy (May 1998). "Resistance of eels (Gymnothorax) to the venom of sea kraits (Laticauda colubrina): a test of coevolution". Toxicon. 36 (4): 619–625. doi:10.1016/S0041-0101(97)00081-0.
- Zimmerman, K. D.; Heatwole, Harold; Davies, H. I. (1 March 1992). "Survival times and resistance to sea snake (Aipysurus laevis) venom by five species of prey fish". Toxicon. 30 (3): 259–264. doi:10.1016/0041-0101(92)90868-6. ISSN 0041-0101.
- Fautin, Daphne G. (1991). "The anemonefish symbiosis: what is known and what is not". Cite journal requires
|journal=(help) - Mebs, Dietrich (15 December 2009). "Chemical biology of the mutualistic relationships of sea anemones with fish and crustaceans". Toxicon. Cnidarian Toxins and Venoms. 54 (8): 1071–1074. doi:10.1016/j.toxicon.2009.02.027. ISSN 0041-0101.
- da Silva, Karen Burke; Nedosyko, Anita (2016), Goffredo, Stefano; Dubinsky, Zvy (eds.), "Sea Anemones and Anemonefish: A Match Made in Heaven", The Cnidaria, Past, Present and Future: The world of Medusa and her sisters, Springer International Publishing, pp. 425–438, doi:10.1007/978-3-319-31305-4_27, ISBN 978-3-319-31305-4, retrieved 2 December 2019
- Nedosyko, Anita M.; Young, Jeanne E.; Edwards, John W.; Silva, Karen Burke da (30 May 2014). "Searching for a Toxic Key to Unlock the Mystery of Anemonefish and Anemone Symbiosis". PLOS ONE. 9 (5): e98449. doi:10.1371/journal.pone.0098449. ISSN 1932-6203. PMC 4039484. PMID 24878777.
- Mebs, D. (1 September 1994). "Anemonefish symbiosis: Vulnerability and resistance of fish to the toxin of the sea anemone". Toxicon. 32 (9): 1059–1068. doi:10.1016/0041-0101(94)90390-5. ISSN 0041-0101.
- Lubbock, R.; Smith, David Cecil (13 February 1980). "Why are clownfishes not stung by sea anemones?". Proceedings of the Royal Society of London. Series B. Biological Sciences. 207 (1166): 35–61. doi:10.1098/rspb.1980.0013.
- Litsios, Glenn; Kostikova, Anna; Salamin, Nicolas (22 November 2014). "Host specialist clownfishes are environmental niche generalists". Proceedings of the Royal Society B: Biological Sciences. 281 (1795): 20133220. doi:10.1098/rspb.2013.3220. PMC 4213602. PMID 25274370.
- Litsios, Glenn; Sims, Carrie A.; Wüest, Rafael O.; Pearman, Peter B.; Zimmermann, Niklaus E.; Salamin, Nicolas (2 November 2012). "Mutualism with sea anemones triggered the adaptive radiation of clownfishes". BMC Evolutionary Biology. 12 (1): 212. doi:10.1186/1471-2148-12-212. ISSN 1471-2148. PMC 3532366. PMID 23122007.
- Conant, Roger, 1909-2003. (1975). A field guide to reptiles and amphibians of Eastern and Central North America ([2d ed.] ed.). Boston,: Houghton Mifflin. ISBN 0-395-19979-4. OCLC 1423604.CS1 maint: extra punctuation (link) CS1 maint: multiple names: authors list (link)
- Holding, Matthew L.; Drabeck, Danielle H.; Jansa, Sharon A.; Gibbs, H. Lisle (November 2016). "Venom Resistance as a Model for Understanding the Molecular Basis of Complex Coevolutionary Adaptations". Integrative and Comparative Biology. 56 (5): 1032–1043. doi:10.1093/icb/icw082. ISSN 1540-7063.
- Weinstein, Scott A.; DeWitt, Clement F.; Smith, Leonard A. (December 1992). "Variability of Venom-Neutralizing Properties of Serum from Snakes of the Colubrid Genus Lampropeltis". Journal of Herpetology. 26 (4): 452. doi:10.2307/1565123.