Nucleotide salvage
A salvage pathway is a pathway in which nucleotides (purine and pyrimidine) are synthesized from intermediates in the degradative pathway for nucleotides.
Salvage pathways are used to recover bases and nucleosides that are formed during degradation of RNA and DNA. This is important in some organs because some tissues cannot undergo de novo synthesis.
The salvaged bases and nucleosides can then be converted back into nucleotides. Salvage pathways are targets for drug development, one family being called antifolates.[1]
Substrates
The salvage pathway requires distinct substrates:
Pyrimidines
Uridine phosphorylase or pyrimidine-nucleoside phosphorylase adds ribose 1-phosphate to the free base uracil, forming uridine. Uridine kinase (aka uridine–cytidine kinase) can then phosphorylate this nucleoside into uridine monophosphate (UMP). UMP/CMP kinase (EC 2.7.4.14) can phosphorylate UMP into uridine diphosphate, which nucleoside diphosphate kinase can phosphorylate into uridine triphosphate.
Thymidine phosphorylase or pyrimidine-nucleoside phosphorylase adds 2-deoxy-alpha-D-ribose 1-phosphate to thymine, forming thymidine. Thymidine kinase can then phosphorylate this compound into thymidine monophosphate (TMP). Thymidylate kinase can phosphorylate TMP into thymidine diphosphate, which nucleoside diphosphate kinase can phosphorylate into thymidine triphosphate.
The nucleosides cytidine and deoxycytidine can be salvaged along the uracil pathway by cytidine deaminase, which converts them to uridine and deoxyuridine, respectively. Alternatively, uridine–cytidine kinase can phosphorylate them into cytidine monophosphate (CMP) or deoxycytidine monophosphate (dCMP). UMP/CMP kinase can phosphorylate (d)CMP into cytidine diphosphate or deoxycytidine diphosphate, which nucleoside diphosphate kinase can phosphorylate into cytidine triphosphate or deoxycytidine triphosphate.
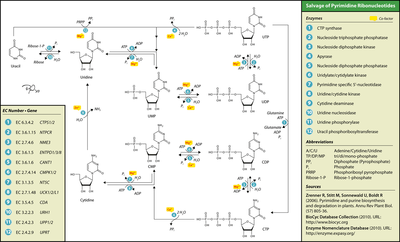
Purines
Phosphoribosyltransferases add activated ribose-5-phosphate (Phosphoribosyl pyrophosphate, PRPP) to bases, creating nucleoside monophosphates. There are two types of phosphoribosyltransferases: adenine phosphoribosyltransferase (APRT) and hypoxanthine-guanine phosphoribosyltransferase (HGPRT). HGPRT is an important enzyme in Purine pathway metabolism and[2] its deficiency is implicated in Lesch-Nyhan syndrome.
The parasite Plasmodium falciparum relies exclusively on the purine salvage pathway for its purine nucleotide requirements.[3][4][5][6] Thus, enzymes constituting the purine salvage pathway in the parasite are potential targets for drug discovery. 5´nucleotidases catalyze the hydrolysis of purine mononucleotides to their respective nucleosides and phosphate.[4][5][6] The nucleosides are taken up in the cell by transporters and are funneled through the salvage pathway. If the nucleoside is adenosine, it is acted upon by adenosine deaminases to convert it into inosine. This metabolite, in turn, is acted upon by purine nucleoside phosphorylase and is converted to hypoxanthine. Hypoxanthine is acted upon by HGXPRT(hypoxanthine guanine xanthine phosphoribosyl transferase) in the parasite to convert the respective nucleobase to its nucleotide monophosphate, respectively (i.e., IMP, GMP or XMP). If it is IMP, this is subsequently acted upon by adenylosuccinate synthase and adenylosuccinate lyase,[3] in a two step process, to convert it into sAMP and AMP, respectively. On the contrary, IMP can also be acted upon by IMP dehydrogenase and GMP synthetase to convert it into GMP.
| Nucleobase | Enzyme | Nucleotide |
| hypoxanthine | hypoxanthine/guanine phosphoribosyl transferase (HGPRT) | IMP |
| guanine | hypoxanthine/guanine phosphoribosyl transferase (HGPRT) | GMP |
| adenine | adenine phosphoribosyltransferase (APRT) | AMP |
Folate biosynthesis
Tetrahydrofolic acid and its derivatives are produced by salvage pathways from GTP.[1]
References
- Kompis IM, Islam K, Then RL (February 2005). "DNA and RNA synthesis: antifolates". Chemical Reviews. 105 (2): 593–620. doi:10.1021/cr0301144. PMID 15700958.
- Ansari MY, Equbal A, Dikhit MR, Mansuri R, Rana S, Ali V, Sahoo GC, Das P (February 2016). "Establishment of correlation between in-silico and in-vitro test analysis against Leishmania HGPRT to inhibitors". International Journal of Biological Macromolecules. 83: 78–96. doi:10.1016/j.ijbiomac.2015.11.051. PMID 26616453.
- Bulusu V, Srinivasan B, Bopanna MP, Balaram H (April 2009). "Elucidation of the substrate specificity, kinetic and catalytic mechanism of adenylosuccinate lyase from Plasmodium falciparum". Biochimica et Biophysica Acta. 1794 (4): 642–54. doi:10.1016/j.bbapap.2008.11.021. PMID 19111634.
- Srinivasan B, Balaram H (2007). "ISN1 nucleotidases and HAD superfamily protein fold: in silico sequence and structure analysis". In Silico Biology. 7 (2): 187–93. PMID 17688444.
- Srinivasan B, Forouhar F, Shukla A, Sampangi C, Kulkarni S, Abashidze M, Seetharaman J, Lew S, Mao L, Acton TB, Xiao R, Everett JK, Montelione GT, Tong L, Balaram H (March 2014). "Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila". The FEBS Journal. 281 (6): 1613–1628. doi:10.1111/febs.12727. PMC 3982195. PMID 24456211.
- Srinivasan B, Kempaiah Nagappa L, Shukla A, Balaram H (April 2015). "Prediction of substrate specificity and preliminary kinetic characterization of the hypothetical protein PVX_123945 from Plasmodium vivax". Experimental Parasitology. 151-152: 56–63. doi:10.1016/j.exppara.2015.01.013. PMID 25655405.