Phage typing
Phage typing is a method used for detecting single strains of bacteria. It is used to trace the source of outbreaks of infections.[1] The viruses that infect bacteria are called bacteriophages ("phages" for short) and some of these can only infect a single strain of bacteria. These phages are used to identify different strains of bacteria within a single species.
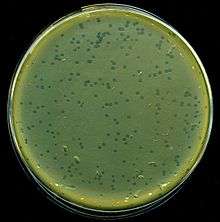
A culture of the strain is grown in the agar and dried. A grid is drawn on the base of the Petri dish to mark out different regions. Inoculation of each square of the grid is done by a different phage. The phage drops are allowed to dry and are incubated: The susceptible phage regions will show a circular clearing where the bacteria have been lysed, and this is used in differentiation.[2] Spotted onto the confluence growth of bacteria
References
- Baggesen DL, Sørensen G, Nielsen EM, Wegener HC. Phage typing of Salmonella Typhimurium - is it still a useful tool for surveillance and outbreak investigation?. Eurosurveillance. 2010;15(4):19471. PMID 20122382.
- Turbadkar SD, Ghadge DP, Patil S, Chowdhary AS, Bharadwaj R (April 2007). "Circulating phage type of Vibrio cholerae in Mumbai". Indian Journal of Medical Microbiology. 25 (2): 177–8. doi:10.4103/0255-0857.32738. PMID 17582202.
[1][2][3][4][5][6][7][8][9][10][11]
- Alamian, Dadar S, Solimani M, Behrozikhah S, Etemadi. 2019. A Case of Identity Confirmation of Brucella abortus S99 by Phage Typing and PCR Methods.
- Cowley LA, Low AS, Pickard D, Boinett CJ, Dallman TJ, Day M, Perry N, Gally DL, Parkhill J, Jenkins C, et al. 2018. Transposon Insertion Sequencing Elucidates Novel Gene Involvement in Susceptibility and Resistance to Phages T4 and T7 in Escherichia coli.
- Kaimal S, D’souza M, Sistla S, Parija SC. 2012. Phage typing in dermatitis cruris pustulosa et atrophicans: does staphylococcal carrier status have a role?
- Lan R, Stevenson G, Donohoe K, Ward L, Reeves PR. 2007. Molecular markers with potential to replace phage typing for Salmonella enterica serovar typhimurium. J Microbiol Methods.
- Lienemann T, Kyyhkynen A, Halkilahti J, Haukka K, Siitonen A. 2015. Characterization of Salmonella Typhimurium isolates from domestically acquired infections in Finland by phage typing, antimicrobial susceptibility testing, PFGE and MLVA. BMC Microbiol.
- Prendergast DM, O’Grady D, Fanning S, Cormican M, Delappe N, Egan J, Mannion C, Fanning J, Gutierrez M. 2011. Application of multiple locus variable number of tandem repeat analysis (MLVA), phage typing and antimicrobial susceptibility testing to subtype Salmonella enterica serovar Typhimurium isolated from pig farms, pork slaughterhouses and meat producing plants in Ireland. Food Microbiol.
- Schürch AC, van Soolingen D. 2012. DNA fingerprinting of Mycobacterium tuberculosis: From phage typing to whole-genome sequencing. Infect Genet Evol.
- Szymczak P, Vogensen FK, Janzen T. 2019. Novel isolates of Streptococcus thermophilus bacteriophages from group 5093 identified with an improved multiplex PCR typing method. Int Dairy J.
- Wiśniewska K, Szewczyk A, Piechowicz L, Bronk M, Samet A, Świeć K. 2012. The use of spa and phage typing for characterization of clinical isolates of methicillin-resistant Staphylococcus aureus in the University Clinical Center in Gdańsk, Poland. Folia Microbiol (Praha).
- Xu D, Zhang J, Liu J, Xu J, Zhou H, Zhang L, Zhu J, Kan B. 2014. Outer Membrane Protein OmpW Is the Receptor for Typing Phage VP5 in the Vibrio cholerae O1 El Tor Biotype. J Virol.
- Ziebell K, Chui L, King R, Johnson S, Boerlin P, Johnson RP. 2017. Subtyping of Canadian isolates of Salmonella Enteritidis using Multiple Locus Variable Number Tandem Repeat Analysis (MLVA) alone and in combination with Pulsed-Field Gel Electrophoresis (PFGE) and phage typing. J Microbiol Methods.