Periplasm
The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria. Using cryo-electron microscopy it has been found that a much smaller periplasmic space is also present in gram-positive bacteria.[1][2]
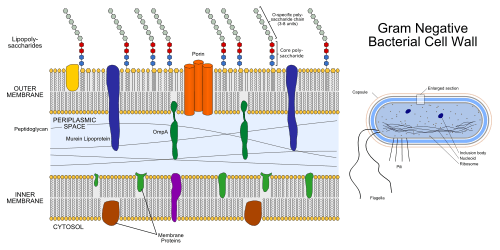
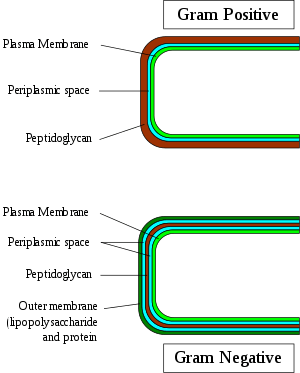
The periplasm may constitute up to 40% of the total cell volume of gram-negative bacteria, but is a much smaller percentage in gram-positive bacteria.[3] Several types of enzyme are present in the periplasm including alkaline phosphatases, cyclic phosphodiesterases, acid phosphatases and 5’-nucleotidases.[4]
Although the bacteria are conventionally divided into two main groups—gram-positive and gram-negative, based upon their Gram-stain retention property—this classification system is ambiguous as it can refer to three distinct aspects (staining result, cell-envelope organization, taxonomic group), which do not necessarily coalesce for some bacterial species.[5][6][7][8] However, although Gram-staining response of bacteria is an empirical criterion, its basis lies in the marked differences in the ultrastructure and chemical composition of the two main kinds of bacteria. These bacteria are distinguished from each other based on the presence or absence of an outer lipid membrane, which is a more reliable and fundamental characteristic of the bacterial cells.[5][9]
All gram-positive bacteria are bounded by a single unit lipid membrane; they generally contain a thick layer (20-80 nm) of peptidoglycan responsible for retaining the Gram-stain. A number of other bacteria which are bounded by a single membrane but stain gram-negative due to either lack of the peptidoglycan layer (viz., mycoplasmas) or their inability to retain the Gram-stain due to their cell wall composition, also show close relationship to the gram-positive bacteria. For the bacterial (prokaryotic) cells that are bounded by a single cell membrane the term "monoderm bacteria" or "monoderm prokaryotes" has been proposed.[5][9] In contrast to gram-positive bacteria, all archetypical gram-negative bacteria are bounded by a cytoplasmic membrane as well as an outer cell membrane; they contain only a thin layer of peptidoglycan (2–3 nm) between these membranes. The presence of both inner and outer cell membranes forms and define the periplasmic space or periplasmic compartment. These bacterial cells with two membranes have been designated as diderm bacteria.[5][9] The distinction between the monoderm and diderm prokaryotes is supported by conserved signature indels in a number of important proteins (viz. DnaK, GroEL).[5][6][9][10]
In diderm bacteria, the periplasm contains a thin cell wall composed of peptidoglycan. In addition, it includes solutes such as ions and proteins, which are involved in wide variety of functions ranging from nutrient binding, transport, folding, degradation, substrate hydrolysis, to peptidoglycan synthesis, electron transport, and alteration of substances toxic to the cell (xenobiotic metabolism).[11] Importantly, the periplasm is devoid of ATP.
References
- , Matias, V. R., and T. J. Beveridge. 2005. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56:240-251. doi:10.1111/j.1365-2958.2005.04535.x.
- , Zuber B, Haenni M, Ribeiro T, Minnig K, Lopes F, Moreillon P, Dubochet J. 2006. Granular layer in the periplasmic space of Gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J Bacteriol. 188:6652-6660. doi:10.1128/JB.00391-06
- Otto Holst; Guntram Seltmann. The Bacterial Cell Wall. Berlin: Springer. ISBN 3-540-42608-6.
- Neu, Harold C.; Heppel, Leon A. (September 1, 1965). "The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts". The Journal of Biological Chemistry. 240 (9): 3685–3692. PMID 4284300.
- Gupta, R.S. (1998) Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria and eukaryotes. Microbiol. Mol. Biol. Rev. 62: 1435-1491.
- Gupta, R.S.(2000) The natural evolutionary relationships among prokaryotes. Crit. Rev. Microbiol. 26: 111-131.
- , Desvaux M, Hébraud M, Talon R, Henderson IR. 2009. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17:139-145. doi:10.1016/j.tim.2009.01.004
- , Sutcliffe IC. 2010. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 18:464-470. doi:10.1016/j.tim.2010.06.005
- Gupta, R. S. (1998). What are archaebacteria: life’s third domain or monoderm prokaryotes related to Gram-positive bacteria? A new proposal for the classification of prokaryotic organisms. Molecular Microbiology.29(3):695-707.
- Gupta, R. S. (2011). Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie van Leeuwenhoek. 100:171-182.
- Klein, Donald W.; Prescott, Lansing M.; Harley, John (2005). Microbiology. Boston: McGraw-Hill Higher Education. ISBN 0-07-295175-3.
Further reading
- D. White, The Physiology and Biochemistry of Prokaryotes, Oxford University Press, Oxford, 2000, pp. 22.