Human parainfluenza viruses
Human parainfluenza viruses (HPIVs) are the viruses that cause human parainfluenza. HPIVs are a paraphyletic group of four distinct single-stranded RNA viruses belonging to the Paramyxoviridae family. These viruses are closely associated with both human and veterinary disease.[2] Virions are approximately 150–250 nm in size and contain negative sense RNA with a genome encompassing about 15,000 nucleotides.[3]
| Human parainfluenza viruses | |
|---|---|
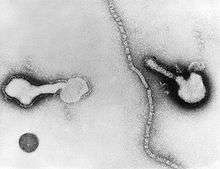 | |
| Transmission electron micrograph of a parainfluenza virus. Two intact particles and free filamentous nucleocapsid | |
| Scientific classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Phylum: | Negarnaviricota |
| Class: | Monjiviricetes |
| Order: | Mononegavirales |
| Family: | Paramyxoviridae |
| Groups included | |
| |
| Cladistically included but traditionally excluded taxa | |
| |
| Human parainfluenza viruses | |
|---|---|
| Specialty | Pulmonology |
The viruses can be detected via cell culture, immunofluorescent microscopy, and PCR.[4] HPIVs remain the second main cause of hospitalisation in children under 5 years of age suffering from a respiratory illness (only Human orthopneumovirus causes more respiratory hospitalisations for this age group).[5]
Classification
The first HPIV was discovered in the late 1950s. The taxonomic division is broadly based on antigenic and genetic characteristics, forming four major serotypes or clades, which today are considered distinct viruses.[6] These include:
| Virus | GenBank acronym | NCBI taxonomy | Notes |
|---|---|---|---|
| Human parainfluenza virus type 1 | HPIV-1 | 12730 | Most common cause of croup |
| Human parainfluenza virus type 2 | HPIV-2 | 11212 | Causes croup and other upper and lower respiratory tract illnesses |
| Human parainfluenza virus type 3 | HPIV-3 | 11216 | Associated with bronchiolitis and pneumonia |
| Human parainfluenza virus type 4 | HPIV-4 | 11203 | Includes subtypes 4a and 4b |
HPIVs belong to two genera: Respirovirus (HPIV-1 & HPIV-3) and Rubulavirus (HPIV-2 & HPIV-4).[3]
Viral structure and organisation
hPIVs are characterised by producing enveloped virions and containing single stranded negative sense RNA.[3] Non-infectious virions have also been reported to contain RNA with positive polarity.[3] HPIV genomes are about 15,000 nucleotides in length and encode six key structural proteins.[3]
The structural gene sequence of HPIVs is as follows: 3′-NP-P-M-F-HN-L-5′ (the protein prefixes and further details are outlined in the table below).[7]
| Structural protein | Location | Function |
|---|---|---|
| Hemagglutinin-neuraminidase (HN) | Envelope | Attachment and cell entry |
| Fusion Protein (F) | Envelope | Fusion and cell entry |
| Matrix Protein (M) | Within the envelope | Assembly |
| Nucleoprotein (NP) | Nucleocapsid | Forms a complex with the RNA genome |
| Phosphoprotein (P) | Nucleocapsid | Forms as part of RNA polymerase complex |
| Large Protein (L) | Nucleocapsid | Forms as part of RNA polymerase complex |
With the advent of reverse genetics, it has been found that the most efficient human parainfluenza viruses (in terms of replication and transcription) have a genome nucleotide total that is divisible by the number 6. This has led to the "rule of six" being coined. Exceptions to this rule have been found and its exact advantages are not fully understood.[8]
Electrophoresis has shown that the molecular weight (MW) of the proteins for the four HPIVs are similar (with the exception of the phosphoprotein, which shows significant variation).[3][9]
Viral entry and replication
Viral replication is initiated only after successful entry into a cell by attachment and fusion between the virus and the host cell lipid membrane. Viral RNA (vRNA) is initially associated with nucleoprotein (NP), phosphoprotein (P) and the large protein (L). The hemagglutinin–neuraminidase (HN) is involved with viral attachment and thus hemadsorption and hemagglutination. Furthermore, the fusion (F) protein is important in aiding the fusion of the host and viral cellular membranes, eventually forming syncytia.[10]
Initially the F protein is in an inactive form (F0) but can be cleaved by proteolysis to form its active form, F1 and F2, linked by di-sulphide bonds. Once complete, this is followed by the HPIV nucleocapsid entering the cytoplasm of the cell. Subsequently, genomic transcription occurs using the viruses own 'viral RNA-dependent RNA polymerase' (L protein). The cell's own ribosomes are then tasked with translation, forming the viral proteins from the viral mRNA.[10]
Towards the end of the process, (after the formation of the viral proteins) the replication of the viral genome occurs. Initially, this occurs with the formation of a positive-sense RNA (intermediate step, necessary for producing progeny), and finally, negative-sense RNA is formed which is then associated with the nucleoprotein. This may then be either packaged and released from the cell by budding or used for subsequent rounds of transcription and replication.[11]
The observable and morphological changes that can be seen in infected cells include the enlargement of the cytoplasm, decreased mitotic activity and 'focal rounding', with the potential formation of multi-nucleate cells (syncytia).[12]
The pathogenicity of HPIVs is mutually dependent on the viruses having the correct accessory proteins that are able to elicit anti-interferon properties. This is a major factor in the clinical significance of disease.[11]
Host range
The main host remains the human. However, infections have been induced in other animals (both under natural and experimental situations), although these were always asymptomatic.[13]
Clinical significance
It is estimated that there are 5 million children with lower respiratory infections (LRI) each year in the United States alone.[14] HPIV-1, HPIV-2 and HPIV-3 have been linked with up to a third of these infections.[15] Upper respiratory infections (URI) are also important in the context of HPIV, however, they are caused to a lesser extent by the virus.[16] The highest rates of serious HPIV illnesses occur among young children, and surveys have shown that about 75% of children aged 5 or older have antibodies to HPIV-1.
For infants and young children, it has been estimated that about 25% will develop "clinically significant disease".[17]
Repeated infection throughout the life of the host is not uncommon and symptoms of later breakouts include upper respiratory tract illness, such as cold and a sore throat.[3] The incubation period for all four serotypes is 1 to 7 days.[18] In immunosuppressed people, parainfluenza virus infections can cause severe pneumonia, which can be fatal.[19]
HPIV-1 and HPIV-2 have been demonstrated to be the principal causative agent behind croup (laryngotracheobronchitis), which is a viral disease of the upper airway and is mainly problematic in children aged 6–48 months of age.[20][21] Biennial epidemics starting in Autumn are associated with both HPIV-1 and 2; however, HPIV-2 can also have yearly outbreaks.[14] Additionally, HPIV-1 tends to cause biennial outbreaks of croup in the Fall. In the United States, large peaks have presently been occurring during odd-numbered years.
HPIV-3 has been closely associated with bronchiolitis and pneumonia and principally targets those aged <1 year.[22]
HPIV-4 remains infrequently detected. However, it is now believed to be more common than previously thought, but is less likely to cause severe disease. By the age of 10, the majority of children are sero-positive for HPIV-4 infection which may be indicative of a large proportion of asymptomatic or mild infections.[3]
Important epidemiological factors that are associated with a higher risk of infection and mortality are those who are immuno-compromised and may be taken ill with more extreme forms of LRI.[13] Associations between HPIVs and neurologic disease are known; for example, hospitalisation with certain HPIVs has a strong association with febrile seizures.[23] HPIV-4B has the strongest association (up to 62%) followed by hPIV-3 and -1.[3]
HPIVs have also been linked with rare cases of virally caused meningitis[24] and Guillain–Barré syndrome.[12]
HPIVs are spread from person to person ('horizontal transmission') by contact with infected secretions through respiratory droplets or contaminated surfaces or objects. Infection can occur when infectious material contacts mucous membranes of the eyes, mouth, or nose, and possibly through the inhalation of droplets generated by a sneeze or cough. HPIVs can remain infectious in airborne droplets for over an hour.
Overall, HPIVs remain best known for its effects on the respiratory system and this appears to be where the majority of the focus has been upon.
Airway inflammation
The inflammation of the airway is a common attribute of HPIV infection. It is believed to occur due to the large scale up-regulation of inflammatory cytokines. Common cytokines, expected to be up-regulated, include IFN–α, various other interleukins (IL–2, IL-6) and TNF–α. Various chemokines and inflammatory proteins are also believed to be associated with the common symptoms of HPIV infection.[12]
Recent evidence seems to suggest that virus-specific antibodies (IgE) may be responsible for mediating large-scale releases of histamine in the trachea, which are believed to cause croup.[12] More detail on the pathways and interactions can be found here.
Immunology
The body's primary defense against HPIV infection remains humoral immunity. This is mainly directed against surface proteins which can be found on the virus. In particular the proteins HN and F prove to be most immunogenic in terms of stimulating the immune system.[12]
Recently the importance of the 'cell mediated immune system' has been scrutinized. Reports have shown that those with defective adaptive immune responses are at a higher risk of severe infection.[12]
Diagnosis
Diagnosis can be made in several ways, encompassing a range of multi-faceted techniques:[4]
- Isolation and detection of the virus in cell culture.
- Detection of viral antigens directly within bodily respiratory tract secretions using immunofluorescence, enzyme immunoassays or fluroimmunoassays.
- Polymerase chain reaction (PCR).
- Analysis of specific IgG antibodies showing a subsequent rise in titre following infection (using paired serum specimens).
Because of the similarity in terms of the antigenic profile between the viruses, hemagglutination assay (HA) or hemadsorption inhibition (HAdI) processes are often used. Both complement fixation, neutralisation and enzyme linked immunosorbent assays – ELISA, can also be used to aid in the process of distinguishing between viral serotypes.[3]
Morbidity and mortality
Mortality caused by HPIVs in developed regions of the world remains rare. Where mortality has occurred, it is principally in the three core risk groups (very young, elderly and immuno-compromised). Long term changes can however be associated with airway remodelling and are believed to be a significant cause of morbidity.[25] The exact associations between HPIVs and diseases such as chronic obstructive pulmonary disease (COPD) are still being investigated.[26]
In developing regions of the world, preschool children remain the highest mortality risk group. Mortality may be a consequence of primary viral infection or secondary problems such as bacterial infection. Predispositions, such as malnutrition and other deficiencies may further elevate the chances of mortality associated with infection.[12]
Overall, LRI's cause approximately 25–30% of total deaths in preschool children in the developing world. HPIVs is believed to be associated with 10% of all LRI cases, thus remaining a significant cause of mortality.[12]
Risk factors
Numerous factors have been suggested and linked to a higher risk of acquiring the infection, inclusive of malnutrition, vitamin A deficiency, absence of breastfeeding during the early stages of life, environmental pollution and overcrowding.[27]
Prevention
Despite decades of research, no vaccines currently exist.[28]
Recombinant technology has however been used to target the formation of vaccines for HPIV-1, -2 and -3 and has taken the form of several live-attenuated intranasal vaccines. Two vaccines in particular were found to be immunogenic and well tolerated against HPIV-3 in phase I trials. HPIV-1 and -2 vaccine candidates remain less advanced.[17]
Vaccine techniques which have been used against HPIVs are not limited to intranasal forms, but also viruses attenuated by cold passage, host range attenuation, chimeric construct vaccines and also introducing mutations with the help of reverse genetics to achieve attenuation.[29]
Maternal antibodies may offer some degree of protection against HPIVs during the early stages of life via the colostrum in breast milk.[30]
Medication
Ribavirin is one medication which has shown good potential for the treatment of HPIV-3 given recent in-vitro tests (in-vivo tests show mixed results).[12] Ribavirin is a broad-spectrum antiviral, and is currently being administered to those who are severely immuno-compromised, despite the lack of conclusive evidence for its benefit.[12] Protein inhibitors and novel forms of medication have also been proposed to relieve the symptoms of infection.[13]
Furthermore, antibiotics may be used if a secondary bacterial infection develops. Corticosteroid treatment and nebulizers are also a first line choice against croup if breathing difficulties ensue.[12]
Interactions with the environment
Parainfluenza viruses last only a few hours in the environment and are inactivated by soap and water. Furthermore, the virus can also be easily destroyed using common hygiene techniques and detergents, disinfectants and antiseptics.[4]
Environmental factors which are important for HPIV survival are pH, humidity, temperature and the medium within which the virus is found. The optimal pH is around the physiologic pH values (7.4 to 8.0), whilst at high temperatures (above 37 °C) and low humidity, infectivity reduces.[31]
The majority of transmission has been linked to close contact, especially in nosocomial infections. Chronic care facilities and doctors' surgeries are also known to be transmission 'hotspots' with transmission occurring via aerosols, large droplets and also fomites (contaminated surfaces).[32]
The exact infectious dose remains unknown.[13]
Economic burden
In the poorest regions of the world, HPIV infection can be measured in terms of mortality. In the developed world where mortality remains rare, the economic costs of the infection can be estimated. Estimates from the US are suggestive of a cost (based on extrapolation) in the region of $200 million per annum.[3]
References
- "Virus Taxonomy: 2018 Release" (html). International Committee on Taxonomy of Viruses (ICTV). October 2018. Retrieved 25 January 2019.
- Vainionpää R, Hyypiä T (April 1994). "Biology of parainfluenza viruses". Clin. Microbiol. Rev. 7 (2): 265–275. doi:10.1128/CMR.7.2.265. PMC 358320. PMID 8055470.
- Henrickson, KJ (April 2003). "Parainfluenza viruses". Clinical Microbiology Reviews. 16 (2): 242–264. doi:10.1128/CMR.16.2.242-264.2003. PMC 153148. PMID 12692097.
- "Human Parainfluenza Viruses". Centers for Disease Control and Prevention (2011). Archived from the original on 20 March 2012. Retrieved 21 March 2012.
- Schmidt, Alexander; Anne Schaap-Nutt; Emmalene J Bartlett; Henrick Schomacker; Jim Boonyaratanakornkit; Ruth A Karron; Peter L Collins (1 February 2011). "Progress in the development of human parainfluenza virus vaccines". Expert Review of Respiratory Medicine. 5 (4): 515–526. doi:10.1586/ers.11.32. PMC 3503243. PMID 21859271.
- "Paramyxoviruses". Parainfluenza Viruses. University of Texas Medical Branch at Galveston. 1996. ISBN 9780963117212. Retrieved 2009-03-15.
- Hunt, Dr. Margaret. "PARAINFLUENZA, RESPIRATORY SYNCYTIAL AND ADENO VIRUSES". Reference.MD. Retrieved 21 March 2012.
- Vulliémoz, D; Roux, L (May 2001). "'Rule of six': how does the Sendai virus RNA polymerase keep count?". Journal of Virology. 75 (10): 4506–4518. doi:10.1128/JVI.75.10.4506-4518.2001. PMC 114204. PMID 11312321.
- Henrickson, K. J (2003). "Parainfluenza Viruses". Clinical Microbiology Reviews. 16 (2): 242–264. doi:10.1128/CMR.16.2.242-264.2003. PMC 153148. PMID 12692097.
- Moscona, A (July 2005). "Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease". The Journal of Clinical Investigation. 115 (7): 1688–1698. doi:10.1172/JCI25669. PMC 1159152. PMID 16007245.
- Chambers R, Takimoto T (2011). Parainfluenza Viruses. eLS. Wiley. doi:10.1002/9780470015902.a0001078.pub3. ISBN 978-0470016176.
- "Parainfluenza Virus: Epidemiology". eMedicine. Retrieved 21 March 2012.
- "HUMAN PARAINFLUENZA VIRUS". Public Health Agency of Canada. 2011-04-19. Retrieved 21 March 2012.
- Henrickson, KJ; Kuhn, SM; Savatski, LL (May 1994). "Epidemiology and cost of infection with human parainfluenza virus types 1 and 2 in young children". Clinical Infectious Diseases. 18 (5): 770–9. doi:10.1093/clinids/18.5.770. PMID 8075269.
- Denny, FW; Clyde WA, Jr (May 1986). "Acute lower respiratory tract infections in nonhospitalized children". The Journal of Pediatrics. 108 (5 Pt 1): 635–46. doi:10.1016/S0022-3476(86)81034-4. PMID 3009769.
- "Acute Respiratory Infections". WHO. Retrieved 21 March 2012.
- Durbin, AP; Karron, RA (December 15, 2003). "Progress in the development of respiratory syncytial virus and parainfluenza virus vaccines". Clinical Infectious Diseases. 37 (12): 1668–1677. doi:10.1086/379775. PMID 14689350.
- "General information: human parainfluenza viruses". Health Protection Agency. Retrieved 21 March 2012.
- Sable CA, Hayden FG (December 1995). "Orthomyxoviral and paramyxoviral infections in transplant patients". Infect. Dis. Clin. North Am. 9 (4): 987–1003. PMID 8747776.
- "CDC - Human Parainfluenza Viruses: Common cold and croup". Archived from the original on 2009-03-03. Retrieved 2009-03-15.
- "Croup Background". Medscape Reference. 2018-10-18. Cite journal requires
|journal=(help) - "Parainfluenza Virus Review". Medscape. Retrieved 21 March 2012.
- Stephen B Greenberg; Robert L Atmar. "Parainfluenza Viruses—New Epidemiology and Vaccine Developments". Touch Infectious Disease. Retrieved 21 March 2012.
- Arguedas, A; Stutman, HR; Blanding, JG (March 1990). "Parainfluenza type 3 meningitis. Report of two cases and review of the literature". Clinical Pediatrics. 29 (3): 175–178. doi:10.1177/000992289002900307. PMID 2155085.
- Dimopoulos, G; Lerikou, M; Tsiodras, S; Chranioti, A; Perros, E; Anagnostopoulou, U; Armaganidis, A; Karakitsos, P (February 2012). "Viral epidemiology of acute exacerbations of chronic obstructive pulmonary disease". Pulmonary Pharmacology & Therapeutics. 25 (1): 12–8. doi:10.1016/j.pupt.2011.08.004. PMID 21983132.
- Beckham, JD; Cadena, A; Lin, J; Piedra, PA; Glezen, WP; Greenberg, SB; Atmar, RL (May 2005). "Respiratory viral infections in patients with chronic, obstructive pulmonary disease". The Journal of Infection. 50 (4): 322–30. doi:10.1016/j.jinf.2004.07.011. PMID 15845430.
- Berman, S (May–Jun 1991). "Epidemiology of acute respiratory infections in children of developing countries". Reviews of Infectious Diseases. 13 Suppl 6: S454–62. doi:10.1093/clinids/13.supplement_6.s454. PMID 1862276.
- Sato M, Wright PF (October 2008). "Current status of vaccines for parainfluenza virus infections". Pediatr. Infect. Dis. J. 27 (10 Suppl): S123–5. doi:10.1097/INF.0b013e318168b76f. PMID 18820572.
- "Parainfluenza Viruses". eLS. Retrieved 21 March 2012.
- "Definition of Human parainfluenza virus". MedicineNet. Retrieved 21 March 2012.
- HAMBLING, MH (December 1964). "Survival of the Respiratory Syncytial Virus During Storage Under Various Conditions". British Journal of Experimental Pathology. 45 (6): 647–55. PMC 2093680. PMID 14245166.
- "Common Cold, Croup and Human Parainfluenza Viruses: Symptoms and Prevention". NewsFlu. Retrieved 21 March 2012.
Further reading
- Henrickson KJ (2003). "Parainfluenza viruses". Clin. Microbiol. Rev. 16 (2): 242–64. doi:10.1128/cmr.16.2.242-264.2003. PMC 153148. PMID 12692097.
- Human Parainfluenza Viruses (HPIVs)
External links
| Classification | |
|---|---|
| External resources |