Orbivirus
The genus Orbivirus is a member of the Reoviridae family, in the subfamily Sedoreovirinae. Unlike the other reoviruses, orbiviruses are arboviruses. This genus currently contains 22 species (including the type species Bluetongue virus) and at least 130 different serotypes.[1][2] Orbiviruses can infect and replicate within a wide range of arthropod and vertebrate hosts. Orbiviruses are named after their characteristic doughnut-shaped capsomers (Orbi in Latin means ring).
| Orbivirus | |
|---|---|
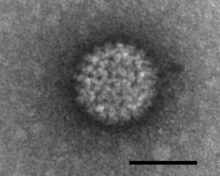 | |
| Negatively stained Bluetongue virus–like particle that caused a cytopathic effect in BHK-21 cells. Scale bar = 50 nm | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Phylum: | incertae sedis |
| Family: | Reoviridae |
| Subfamily: | Sedoreovirinae |
| Genus: | Orbivirus |
| Type species | |
| Bluetongue virus | |
| Species | |
| |
Many orbiviruses are transmitted by ticks or haematophagus insect vectors (Culicoides, mosquitoes and sand flies) and have a wide host range that includes cattle, goats and sheep, wild ruminants, equids, camelids, marsupials, sloths, bats, birds, large canine and feline carnivores, and humans.
The three economically most important orbiviruses are Bluetongue virus, African horse sickness virus, and Epizootic hemorrhagic disease virus, all of which are transmitted by Culicoides species.
Virion
The virons are nonenveloped particles that are 70–80 nm in diameter. The virus particles are spherical in appearance and have icosahedral symmetry.[3] An outer and an inner capsid layer surround the genome, and have T=13 and T=2 symmetry, respectively.[2] The viron is constructed of two concentric protein shells, the subcore layer which contain 120 copies/particle of the VP3 and the core-surface layer composed of 780 copies/particle of the VP7. VP1, VP4, and VP6 are minor enzymatic proteins that are packaged along with the 10 genome segments within the central space of the virus core. The orbivirus outer-capsid layer is composed of two additional structural proteins (VP2 and VP5) which mediate cell-attachment and penetration during initiation of infection. The outer-capsid proteins are more variable than the core proteins and most of the non-structural proteins and the specificity of their reactions with neutralising antibodies determines the virus serotype.
Genome
These viruses have double-stranded RNA genomes, so are classified as Class III viruses. Their genome is linear and is segmented into 10 segments of various lengths. One copy of each gene segment is packaged per virion. In most cases, each gene segment encodes a single open reading frame (ORF). The genome encodes seven major structural proteins (VP1–VP7) and three major nonstructural proteins (NS1-NS3). Exceptions to the one gene-one protein rule are segment 9 (Seg-9) and segment 10 (Seg-10), both of which encode two nearly identical proteins initiated from in-phase AUG codons close together near the upstream termini (VP6 and VP6a encoded by Seg-9: NS3 and NS3a encoded by Seg-10)
An ORF spans almost the entire length of genome segment 9 and encodes VP6 (the viral helicase). A second ORF (OrfX) is also present on this segment and encodes a fourth nonstructural protein (NS4), which was predicted from sequence analysis of various orbiviruses including segment 9 of Great Island virus which contained a long NS4 ORF (around 21kDa). The existence of NS4 was experimentally confirmed in both insect-borne and tick-borne orbiviruses in 2011.[4]
NS1 is the most abundant protein in BTV infected cells. It forms tubules that may be involved in translocation of progeny virus particles to the cell membrane. NS2 is phosphorylated by cellular kinases and is an important matrix protein of the granular viral inclusion bodies that form within the cytoplasm of infected cells. These viral inclusion bodies act as the centres of viral replication. The membrane glycoproteins NS3 and NS3a are expressed in large numbers in insect cells, but not in mammalian cells. They are involved in the release of progeny virus particles from infected cells and may be involved in determination of both vector competence and virulence.
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Orbivirus | Icosahedral | T=13, T=2 | Non-enveloped | Linear | Segmented |
Replication
Many orbiviruses preferentially infect vascular endothelial cells. Orbiviruses enter the host cell by endocytosis and the outer capsid is subsequently removed. The whole cycle of viral replication takes place within the cytoplasm of the host cell. Transcription of the viral genome into mRNA occurs within the core particle and mRNA is translated into proteins using the host cell ribosomes. Viral proteins are synthesized 2–14 days after initial infection. New virons self-assemble within the cytoplasm and are then released from the host cell by budding. During the budding process, they transiently acquire a lipid envelope which can be detected for a short period of time following their release, but this is subsequently lost.
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Orbivirus | Vertebrates; mosquitoes; midges; gnats; sandflies; ticks | vascular endothelial cells | Cell receptor endocytosis | Cell death | Cytoplasm | Cytoplasm | Arthropod bite |
Pathogenesis
Orbiviruses primarily cause diseases in animals. The different Orbivirus species have different host specificities. Orbiviruses are vector-borne pathogens transmitted between vertebrate hosts by vectors such as mosquitoes, midges, gnats, sandflies, and ticks. Bluetongue virus (BTV) is an Orbivirus that causes bluetongue disease in sheep, cattle, goats, and wild ungulates. BTV has been in the forefront of molecular studies for last three decades and now represents one of the best understood viruses at the molecular and structural levels.[5][6] Other species of orbiviruses are responsible for other diseases of animals such as African horse sickness and equine encephalosis virus.[7]
Classification
ICTV taxonomy
Group: dsRNA
- Family: Reoviridae
- Sub-Family: Sedoreovirinae
- Genus: Orbivirus
- African horse sickness virus
- Bluetongue virus
- Changuinola virus
- Chenuda virus
- Chobar Gorge virus
- Corriparta virus
- Epizootic hemorrhagic disease virus
- Equine encephalosis virus
- Eubenangee virus
- Great Island virus
- Ieri virus
- Lebombo virus
- Orungo virus
- Palyam virus
- Peruvian horse sickness virus
- St Croix River virus
- Umatilla virus
- Wad Medani virus
- Wallal virus
- Warrego virus
- Wongorr virus
- Yunnan orbivirus
Serogroups
This genus has been divided into several (at least 14) serogroups.[8][9] The serogroups have been divided in some cases into subgroups. A number of these viruses have yet to be assigned to a serogroup. The serogroups are differentiated on the basis of a fourfold or greater difference in antibody based tests. These tests include ELISAs and complement fixation tests.
- Lebombo virus
- Pata virus
- African horse sickness virus (Serotypes 1–9)
- African horse sickness virus
- Bluetongue virus (serotypes 1–26)
- Changuinola serogroup
- Altamira virus
- Almeirim virus
- Caninde virus
- Changuinola virus
- Irituia virus
- Jamanxi virus
- Jari virus
- Gurupi virus
- Monte Dourado virus
- Ourem virus
- Purus virus
- Saraca virus
- Corriparta serogroup
- Acado virus
- Corriparta virus
- Eubenangee serogroup
- Eubenangee virus
- Tilligerry virus
- Epizootic haemorrhagic disease serogroup
- Epizootic hemorrhagic disease virus 1
- Epizootic hemorrhagic disease virus 2
- Kawanabe virus
- Equine encephalosis serogroup
- Equine encephalosis virus
- Great Island virus
- Kemerovo virus
- Essaouira virus
- Kala iris virus
- Mill Door/79 virus
- Rabbit syncytium virus
- Tribeč virus
- Broadhaven virus
- Chenuda subgroup
- Wad Medani subgroup
- Orungo virus(serotypes 1–3)
- Palyam serogroup
- Abadina virus
- Apies River virus
- Bunyip Creek virus
- Chuzan (Kasba) virus
- CSIRO Village virus
- D'Aguilar virus
- Marrakai virus
- Petevo virus
- Vellore virus
- Umatilla serogroup
- Llano Seco virus
- Minnal virus
- Netivot virus
- Umatilla virus
- Wallal serogroup
- Wallal virus
- Warrego serogroup
- Mitchell River virus
- Warrego virus
- Wongorr serogroup
- Paroo River virus
- Picola virus
- Wongorr virus
Species
There are currently 22 recognized virus species as well as 13 unassigned viruses in this genus
Species (with constituent viruses):
- African horse sickness virus
- Bluetongue virus
- Chuzan virus
- Corriparta virus
- Epizootic haemorrhagic disease virus
- Equine encephalosis virus
- Eubenengee virus
- Eubenengee virus
- Ngoupe virus
- Tilligerry virus
- Great Island virus
- Broadhaven virus
- Great Island virus
- Kemerovo virus
- Lipovnik virus
- Tribec viruses
- Lake Clarendon virus
- Palyam virus
- CSIRO Village virus
- Pata virus
- Peruvian horse sickness virus
- Elsey virus
- St Croix river virus
- Umatilla virus
- Stretch Lagoon virus
- Umatilla virus
- Wallal virus
- Warrego virus
- Wongorr virus
- Yunnan virus
- Middle Point virus
Vector groups
These viruses are known to be transmitted by midges (Culicoides), mosquitoes, and ticks. Those viruses transmitted by a particular type of vector seem to be related both genetically and serologically.
Midge vector group
- African horse sickness virus
- Bluetongue virus
- Chuzan virus
- Epizootic haemorrhagic disease virus
- Equine encephalosis virus
- Eubenengee virus
- Palyam virus
- Wallal virus
- Warrego virus
Mosquito vector group
- Corriparta virus
- Peruvian horse sickness virus
- Wongorr virus
- Umatilla virus
- Yunnan virus
Tick vector group
- Broadhaven virus
- Great Island virus
- Kemerovo virus
- Lipovnik virus
- Tribec viruses
Undetermined vector
The vector(s) of the St Croix river virus are not known and based on its genome sequence this virus does not appear to group with any other vector group.
Notes
The tick group may be ancestral to the other groups.[10]
History
In 1719, African horse sickness virus caused the first major recorded orbivirus epidemic, killing 1,500 animals. The most historically significant outbreak of orbivirus occurred in 1854–1855, when AHSV infected 70,000 horses. AHSV was discovered to be a virus in 1900 and bluetongue disease followed shortly thereafter in 1905. Outbreaks have occurred sporadically in the 20th and 21st centuries.[3]
See also
- Animal viruses
See also
- emedicine Orbivirus
- Draft of the Orbivirus genus description for the ICTV seventh report
- Mertens, (2001) Orbiviruses and Bluetongue virus. In: Encyclopedia of Life Sciences. John Wiley & Sons, Ltd: Chichester. www.els.net doi:10.1038/npg.els.0001010
References
- ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- "Viral Zone". ExPASy. Retrieved 15 June 2015.
- Knipe, David M. (17 June 2013). Fields Virology. Wolters Kluwer Health. ISBN 9781451105636.
- Belhouchet M, Mohd Jaafar F, Firth AE, Grimes JM, Mertens PP, Attoui H (2011) Detection of a fourth orbivirus non-structural protein. PLoS One 6(10):e25697.
- Roy P (2008). "Molecular Dissection of Bluetongue Virus". Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6.
- Roy P (2008). "Structure and Function of Bluetongue Virus and its Proteins". Segmented Double-stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-21-9.
- Viljoen and Huismans (1989) The Characterization of Equine Encephalosis Virus and the Development of Genomic Probes. J. gen. Virol. 70, 2007–2015.
- Dilcher M, Hasib L, Lechner M, Wieseke N, Middendorf M, Marz M, Koch A, Spiegel M, Dobler G, Hufert FT, Weidmann M (2000) Genetic characterization of Tribeč virus and Kemerovo virus, two tick-transmitted human-pathogenic Orbiviruses. Virology
- Warrell DA, Cox TM, Firth JD (2003) Oxford textbook of Medicine. Oxford
- Belaganahalli MN, Maan S, Maan NS, Tesh R, Attoui H, Mertens PP (2011) Umatilla virus genome sequencing and phylogenetic analysis: identification of stretch lagoon orbivirus as a new member of the Umatilla virus species. PLoS One 6(8):e23605.