Glycolysis
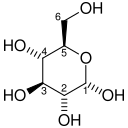
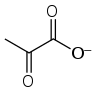
Glycolysis (from glycose, an older term[1] for glucose + -lysis degradation) is the metabolic pathway that converts glucose C6H12O6, into pyruvate, CH3COCOO−, and a hydrogen ion, H+. The free energy released in this process is used to form the high-energy molecules ATP (adenosine triphosphate) and NADH (reduced nicotinamide adenine dinucleotide).[2][3] Glycolysis is a sequence of ten enzyme-catalyzed reactions. Most monosaccharides, such as fructose and galactose, can be converted to one of these intermediates. The intermediates may also be directly useful rather than just utilized as steps in the overall reaction. For example, the intermediate dihydroxyacetone phosphate (DHAP) is a source of the glycerol that combines with fatty acids to form fat.
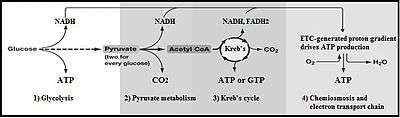
Glycolysis is an oxygen-independent metabolic pathway. The wide occurrence of glycolysis indicates that it is an ancient metabolic pathway.[4] Indeed, the reactions that constitute glycolysis and its parallel pathway, the pentose phosphate pathway, occur metal-catalyzed under the oxygen-free conditions of the Archean oceans, also in the absence of enzymes.[5]
In most organisms, glycolysis occurs in the cytosol. The most common type of glycolysis is the Embden–Meyerhof–Parnas (EMP pathway), which was discovered by Gustav Embden, Otto Meyerhof, and Jakub Karol Parnas. Glycolysis also refers to other pathways, such as the Entner–Doudoroff pathway and various heterofermentative and homofermentative pathways. However, the discussion here will be limited to the Embden–Meyerhof–Parnas pathway.[6]
The glycolysis pathway can be separated into two phases:[2]
- The Preparatory (or Investment) Phase – wherein ATP is consumed.
- The Pay Off Phase – wherein ATP is produced.
Overview
The overall reaction of glycolysis is:
The use of symbols in this equation makes it appear unbalanced with respect to oxygen atoms, hydrogen atoms, and charges. Atom balance is maintained by the two phosphate (Pi) groups:[7]
- Each exists in the form of a hydrogen phosphate anion (HPO42−), dissociating to contribute 2 H+ overall
- Each liberates an oxygen atom when it binds to an ADP (adenosine diphosphate) molecule, contributing 2 O overall
Charges are balanced by the difference between ADP and ATP. In the cellular environment, all three hydroxyl groups of ADP dissociate into −O− and H+, giving ADP3−, and this ion tends to exist in an ionic bond with Mg2+, giving ADPMg−. ATP behaves identically except that it has four hydroxyl groups, giving ATPMg2−. When these differences along with the true charges on the two phosphate groups are considered together, the net charges of −4 on each side are balanced.
For simple fermentations, the metabolism of one molecule of glucose to two molecules of pyruvate has a net yield of two molecules of ATP. Most cells will then carry out further reactions to 'repay' the used NAD+ and produce a final product of ethanol or lactic acid. Many bacteria use inorganic compounds as hydrogen acceptors to regenerate the NAD+.
Cells performing aerobic respiration synthesize much more ATP, but not as part of glycolysis. These further aerobic reactions use pyruvate and NADH + H+ from glycolysis. Eukaryotic aerobic respiration produces approximately 34 additional molecules of ATP for each glucose molecule, however most of these are produced by a mechanism vastly different than the substrate-level phosphorylation in glycolysis.
The lower-energy production, per glucose, of anaerobic respiration relative to aerobic respiration, results in greater flux through the pathway under hypoxic (low-oxygen) conditions, unless alternative sources of anaerobically oxidizable substrates, such as fatty acids, are found.
| Metabolism of common monosaccharides, including glycolysis, gluconeogenesis, glycogenesis and glycogenolysis |
|---|
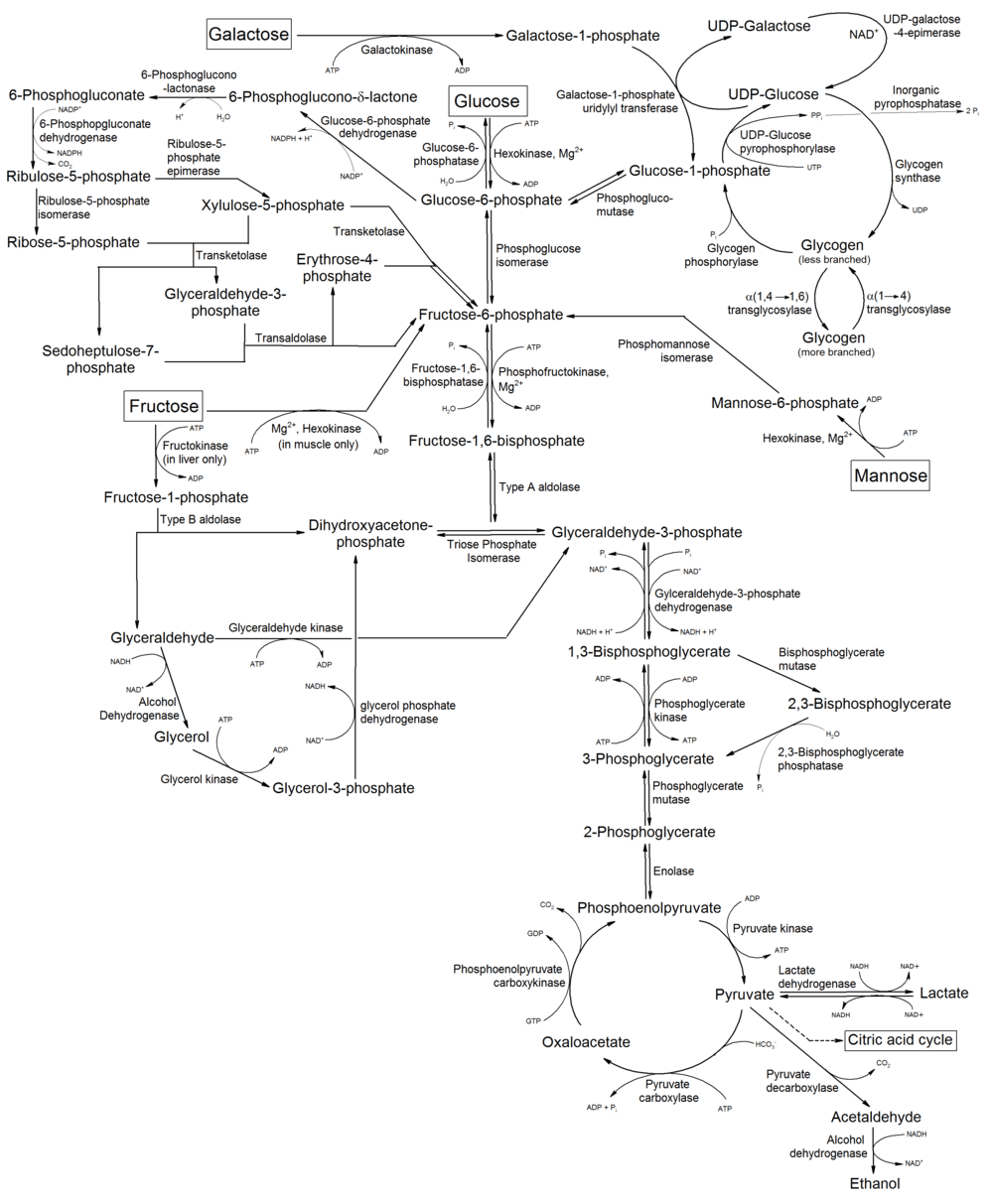 |
History
The pathway of glycolysis as it is known today took almost 100 years to fully discover.[8] The combined results of many smaller experiments were required in order to understand the pathway as a whole.
The first steps in understanding glycolysis began in the nineteenth century with the wine industry. For economic reasons, the French wine industry sought to investigate why wine sometimes turned distasteful, instead of fermenting into alcohol. French scientist Louis Pasteur researched this issue during the 1850s, and the results of his experiments began the long road to elucidating the pathway of glycolysis.[9] His experiments showed that fermentation occurs by the action of living microorganisms; and that yeast's glucose consumption decreased under aerobic conditions of fermentation, in comparison to anaerobic conditions (the Pasteur effect).[10]

Insight into the component steps of glycolysis were provided by the non-cellular fermentation experiments of Eduard Buchner during the 1890s.[11][12] Buchner demonstrated that the conversion of glucose to ethanol was possible using a non-living extract of yeast (due to the action of enzymes in the extract).[13] This experiment not only revolutionized biochemistry, but also allowed later scientists to analyze this pathway in a more controlled lab setting. In a series of experiments (1905-1911), scientists Arthur Harden and William Young discovered more pieces of glycolysis.[14] They discovered the regulatory effects of ATP on glucose consumption during alcohol fermentation. They also shed light on the role of one compound as a glycolysis intermediate: fructose 1,6-bisphosphate.[15]
The elucidation of fructose 1,6-bisphosphate was accomplished by measuring CO2 levels when yeast juice was incubated with glucose. CO2 production increased rapidly then slowed down. Harden and Young noted that this process would restart if an inorganic phosphate (Pi) was added to the mixture. Harden and Young deduced that this process produced organic phosphate esters, and further experiments allowed them to extract fructose diphosphate (F-1,6-DP).
Arthur Harden and William Young along with Nick Sheppard determined, in a second experiment, that a heat-sensitive high-molecular-weight subcellular fraction (the enzymes) and a heat-insensitive low-molecular-weight cytoplasm fraction (ADP, ATP and NAD+ and other cofactors) are required together for fermentation to proceed. This experiment begun by observing that dialyzed (purified) yeast juice could not ferment or even create a sugar phosphate. This mixture was rescued with the addition of undialyzed yeast extract that had been boiled. Boiling the yeast extract renders all proteins inactive (as it denatures them). The ability of boiled extract plus dialyzed juice to complete fermentation suggests that the cofactors were non-protein in character.[14]

In the 1920s Otto Meyerhof was able to link together some of the many individual pieces of glycolysis discovered by Buchner, Harden, and Young. Meyerhof and his team were able to extract different glycolytic enzymes from muscle tissue, and combine them to artificially create the pathway from glycogen to lactic acid.[16][17]
In one paper, Meyerhof and scientist Renate Junowicz-Kockolaty investigated the reaction that splits fructose 1,6-diphosphate into the two triose phosphates. Previous work proposed that the split occurred via 1,3-diphosphoglyceraldehyde plus an oxidizing enzyme and cozymase. Meyerhoff and Junowicz found that the equilibrium constant for the isomerase and aldoses reaction were not affected by inorganic phosphates or any other cozymase or oxidizing enzymes. They further removed diphosphoglyceraldehyde as a possible intermediate in glycolysis.[17]
With all of these pieces available by the 1930s, Gustav Embden proposed a detailed, step-by-step outline of that pathway we now know as glycolysis.[18] The biggest difficulties in determining the intricacies of the pathway were due to the very short lifetime and low steady-state concentrations of the intermediates of the fast glycolytic reactions. By the 1940s, Meyerhof, Embden and many other biochemists had finally completed the puzzle of glycolysis.[17] The understanding of the isolated pathway has been expanded in the subsequent decades, to include further details of its regulation and integration with other metabolic pathways.
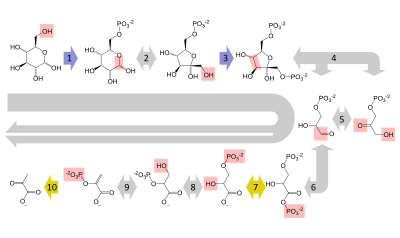
Sequence of reactions
Summary of reactions
Hexokinase
Glucose 6-phosphate
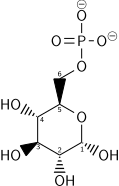
Glucose-6-phosphate
isomerase
Fructose 6-phosphate

phosphofructokinase-1
Fructose 1,6-bisphosphate
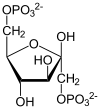
Fructose-bisphosphate
aldolase
Dihydroxyacetone phosphate
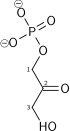
+
Glyceraldehyde 3-phosphate
Triosephosphate
isomerase
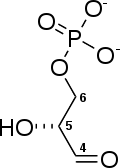
2 × Glyceraldehyde 3-phosphate

Glyceraldehyde-3-phosphate
dehydrogenase
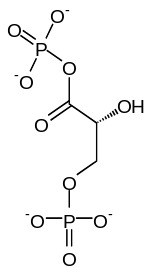
Phosphoglycerate kinase
2 × 3-Phosphoglycerate
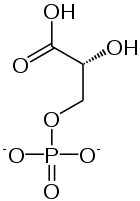
Phosphoglycerate mutase
2 × 2-Phosphoglycerate
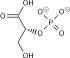
Phosphopyruvate
hydratase (Enolase)
2 × Phosphoenolpyruvate
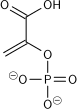
Pyruvate kinase
2 × Pyruvate
Preparatory phase
The first five steps are regarded as the preparatory (or investment) phase, since they consume energy to convert the glucose into two three-carbon sugar phosphates[2] (G3P).
|
||||||||||||||||||||
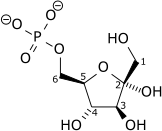
The first step in glycolysis is phosphorylation of glucose by a family of enzymes called hexokinases to form glucose 6-phosphate (G6P). This reaction consumes ATP, but it acts to keep the glucose concentration low, promoting continuous transport of glucose into the cell through the plasma membrane transporters. In addition, it blocks the glucose from leaking out – the cell lacks transporters for G6P, and free diffusion out of the cell is prevented due to the charged nature of G6P. Glucose may alternatively be formed from the phosphorolysis or hydrolysis of intracellular starch or glycogen.
In animals, an isozyme of hexokinase called glucokinase is also used in the liver, which has a much lower affinity for glucose (Km in the vicinity of normal glycemia), and differs in regulatory properties. The different substrate affinity and alternate regulation of this enzyme are a reflection of the role of the liver in maintaining blood sugar levels.
Cofactors: Mg2+
|
||||||||||||||||||||
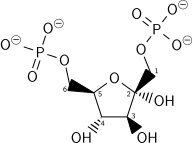
G6P is then rearranged into fructose 6-phosphate (F6P) by glucose phosphate isomerase. Fructose can also enter the glycolytic pathway by phosphorylation at this point.
The change in structure is an isomerization, in which the G6P has been converted to F6P. The reaction requires an enzyme, phosphoglucose isomerase, to proceed. This reaction is freely reversible under normal cell conditions. However, it is often driven forward because of a low concentration of F6P, which is constantly consumed during the next step of glycolysis. Under conditions of high F6P concentration, this reaction readily runs in reverse. This phenomenon can be explained through Le Chatelier's Principle. Isomerization to a keto sugar is necessary for carbanion stabilization in the fourth reaction step (below).
|
||||||||||||||||||||
The energy expenditure of another ATP in this step is justified in 2 ways: The glycolytic process (up to this step) becomes irreversible, and the energy supplied destabilizes the molecule. Because the reaction catalyzed by Phosphofructokinase 1 (PFK-1) is coupled to the hydrolysis of ATP (an energetically favorable step) it is, in essence, irreversible, and a different pathway must be used to do the reverse conversion during gluconeogenesis. This makes the reaction a key regulatory point (see below). This is also the rate-limiting step.
Furthermore, the second phosphorylation event is necessary to allow the formation of two charged groups (rather than only one) in the subsequent step of glycolysis, ensuring the prevention of free diffusion of substrates out of the cell.
The same reaction can also be catalyzed by pyrophosphate-dependent phosphofructokinase (PFP or PPi-PFK), which is found in most plants, some bacteria, archea, and protists, but not in animals. This enzyme uses pyrophosphate (PPi) as a phosphate donor instead of ATP. It is a reversible reaction, increasing the flexibility of glycolytic metabolism.[19] A rarer ADP-dependent PFK enzyme variant has been identified in archaean species.[20]
Cofactors: Mg2+
|
||||||||||||||||||||||||||
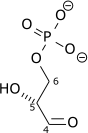
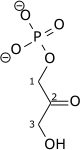
Destabilizing the molecule in the previous reaction allows the hexose ring to be split by aldolase into two triose sugars: dihydroxyacetone phosphate (a ketose), and glyceraldehyde 3-phosphate (an aldose). There are two classes of aldolases: class I aldolases, present in animals and plants, and class II aldolases, present in fungi and bacteria; the two classes use different mechanisms in cleaving the ketose ring.
Electrons delocalized in the carbon-carbon bond cleavage associate with the alcohol group. The resulting carbanion is stabilized by the structure of the carbanion itself via resonance charge distribution and by the presence of a charged ion prosthetic group.
|
||||||||||||||||||||
Triosephosphate isomerase rapidly interconverts dihydroxyacetone phosphate with glyceraldehyde 3-phosphate (GADP) that proceeds further into glycolysis. This is advantageous, as it directs dihydroxyacetone phosphate down the same pathway as glyceraldehyde 3-phosphate, simplifying regulation.
Pay-off phase
The second half of glycolysis is known as the pay-off phase, characterised by a net gain of the energy-rich molecules ATP and NADH.[2] Since glucose leads to two triose sugars in the preparatory phase, each reaction in the pay-off phase occurs twice per glucose molecule. This yields 2 NADH molecules and 4 ATP molecules, leading to a net gain of 2 NADH molecules and 2 ATP molecules from the glycolytic pathway per glucose.
|
||||||||||||||||||||
The aldehyde groups of the triose sugars are oxidised, and inorganic phosphate is added to them, forming 1,3-bisphosphoglycerate.
The hydrogen is used to reduce two molecules of NAD+, a hydrogen carrier, to give NADH + H+ for each triose.
Hydrogen atom balance and charge balance are both maintained because the phosphate (Pi) group actually exists in the form of a hydrogen phosphate anion (HPO42−),[7] which dissociates to contribute the extra H+ ion and gives a net charge of -3 on both sides.
Here, Arsenate (AsO43−), an anion akin to inorganic phosphate may replace phosphate as a substrate to form 1-arseno-3-phosphoglycerate. This, however, is unstable and readily hydrolyzes to form 3-phosphoglycerate, the intermediate in the next step of the pathway. As a consequence of bypassing this step, the molecule of ATP generated from 1-3 bisphosphoglycerate in the next reaction will not be made, even though the reaction proceeds. As a result, arsenate is an uncoupler of glycolysis.[21]
|
||||||||||||||||||||
This step is the enzymatic transfer of a phosphate group from 1,3-bisphosphoglycerate to ADP by phosphoglycerate kinase, forming ATP and 3-phosphoglycerate. At this step, glycolysis has reached the break-even point: 2 molecules of ATP were consumed, and 2 new molecules have now been synthesized. This step, one of the two substrate-level phosphorylation steps, requires ADP; thus, when the cell has plenty of ATP (and little ADP), this reaction does not occur. Because ATP decays relatively quickly when it is not metabolized, this is an important regulatory point in the glycolytic pathway.
ADP actually exists as ADPMg−, and ATP as ATPMg2−, balancing the charges at -5 both sides.
Cofactors: Mg2+
|
||||||||||||||||||||
Phosphoglycerate mutase isomerises 3-phosphoglycerate into 2-phosphoglycerate.
|
||||||||||||||||||||
Enolase next converts 2-phosphoglycerate to phosphoenolpyruvate. This reaction is an elimination reaction involving an E1cB mechanism.
Cofactors: 2 Mg2+: one "conformational" ion to coordinate with the carboxylate group of the substrate, and one "catalytic" ion that participates in the dehydration.
|
||||||||||||||||||||
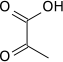
A final substrate-level phosphorylation now forms a molecule of pyruvate and a molecule of ATP by means of the enzyme pyruvate kinase. This serves as an additional regulatory step, similar to the phosphoglycerate kinase step.
Cofactors: Mg2+
Biochemical logic
The existence of more than one point of regulation indicates that intermediates between those points enter and leave the glycolysis pathway by other processes. For example, in the first regulated step, hexokinase converts glucose into glucose-6-phosphate. Instead of continuing through the glycolysis pathway, this intermediate can be converted into glucose storage molecules, such as glycogen or starch. The reverse reaction, breaking down, e.g., glycogen, produces mainly glucose-6-phosphate; very little free glucose is formed in the reaction. The glucose-6-phosphate so produced can enter glycolysis after the first control point.
In the second regulated step (the third step of glycolysis), phosphofructokinase converts fructose-6-phosphate into fructose-1,6-bisphosphate, which then is converted into glyceraldehyde-3-phosphate and dihydroxyacetone phosphate. The dihydroxyacetone phosphate can be removed from glycolysis by conversion into glycerol-3-phosphate, which can be used to form triglycerides.[22] Conversely, triglycerides can be broken down into fatty acids and glycerol; the latter, in turn, can be converted into dihydroxyacetone phosphate, which can enter glycolysis after the second control point.
Free energy changes
| Compound | Concentration / mM |
|---|---|
| Glucose | 5.0 |
| Glucose-6-phosphate | 0.083 |
| Fructose-6-phosphate | 0.014 |
| Fructose-1,6-bisphosphate | 0.031 |
| Dihydroxyacetone phosphate | 0.14 |
| Glyceraldehyde-3-phosphate | 0.019 |
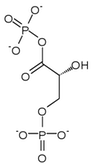
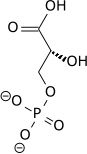
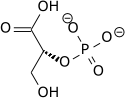
| 1,3-Bisphosphoglycerate | 0.001 |
| 2,3-Bisphosphoglycerate | 4.0 |
| 3-Phosphoglycerate | 0.12 |
| 2-Phosphoglycerate | 0.03 |
| Phosphoenolpyruvate | 0.023 |
| Pyruvate | 0.051 |
| ATP | 1.85 |
| ADP | 0.14 |
| Pi | 1.0 |
The change in free energy, ΔG, for each step in the glycolysis pathway can be calculated using ΔG = ΔG°' + RTln Q, where Q is the reaction quotient. This requires knowing the concentrations of the metabolites. All of these values are available for erythrocytes, with the exception of the concentrations of NAD+ and NADH. The ratio of NAD+ to NADH in the cytoplasm is approximately 1000, which makes the oxidation of glyceraldehyde-3-phosphate (step 6) more favourable.
Using the measured concentrations of each step, and the standard free energy changes, the actual free energy change can be calculated. (Neglecting this is very common - the delta G of ATP hydrolysis in cells is not the standard free energy change of ATP hydrolysis quoted in textbooks).
| Step | Reaction | ΔG°' / (kJ/mol) | ΔG / (kJ/mol) |
|---|---|---|---|
| 1 | Glucose + ATP4− → Glucose-6-phosphate2− + ADP3− + H+ | -16.7 | -34 |
| 2 | Glucose-6-phosphate2− → Fructose-6-phosphate2− | 1.67 | -2.9 |
| 3 | Fructose-6-phosphate2− + ATP4− → Fructose-1,6-bisphosphate4− + ADP3− + H+ | -14.2 | -19 |
| 4 | Fructose-1,6-bisphosphate4− → Dihydroxyacetone phosphate2− + Glyceraldehyde-3-phosphate2− | 23.9 | -0.23 |
| 5 | Dihydroxyacetone phosphate2− → Glyceraldehyde-3-phosphate2− | 7.56 | 2.4 |
| 6 | Glyceraldehyde-3-phosphate2− + Pi2− + NAD+ → 1,3-Bisphosphoglycerate4− + NADH + H+ | 6.30 | -1.29 |
| 7 | 1,3-Bisphosphoglycerate4− + ADP3− → 3-Phosphoglycerate3− + ATP4− | -18.9 | 0.09 |
| 8 | 3-Phosphoglycerate3− → 2-Phosphoglycerate3− | 4.4 | 0.83 |
| 9 | 2-Phosphoglycerate3− → Phosphoenolpyruvate3− + H2O | 1.8 | 1.1 |
| 10 | Phosphoenolpyruvate3− + ADP3− + H+ → Pyruvate− + ATP4− | -31.7 | -23.0 |
From measuring the physiological concentrations of metabolites in an erythrocyte it seems that about seven of the steps in glycolysis are in equilibrium for that cell type. Three of the steps — the ones with large negative free energy changes — are not in equilibrium and are referred to as irreversible; such steps are often subject to regulation.
Step 5 in the figure is shown behind the other steps, because that step is a side-reaction that can decrease or increase the concentration of the intermediate glyceraldehyde-3-phosphate. That compound is converted to dihydroxyacetone phosphate by the enzyme triose phosphate isomerase, which is a catalytically perfect enzyme; its rate is so fast that the reaction can be assumed to be in equilibrium. The fact that ΔG is not zero indicates that the actual concentrations in the erythrocyte are not accurately known.
Regulation of the rate limiting enzymes
The four regulatory enzymes are hexokinase (or glucokinase in the liver), phosphofructokinase, and pyruvate kinase. The flux through the glycolytic pathway is adjusted in response to conditions both inside and outside the cell. The internal factors that regulate glycolysis do so primarily to provide ATP in adequate quantities for the cell’s needs. The external factors act primarily on the liver, fat tissue, and muscles, which can remove large quantities of glucose from the blood after meals (thus preventing hyperglycemia by storing the excess glucose as fat or glycogen, depending on the tissue type). The liver is also capable of releasing glucose into the blood between meals, during fasting, and exercise thus preventing hypoglycemia by means of glycogenolysis and gluconeogenesis. These latter reactions coincide with the halting of glycolysis in the liver.
In animals, regulation of blood glucose levels by the pancreas in conjunction with the liver is a vital part of homeostasis. The beta cells in the pancreatic islets are sensitive to the blood glucose concentration.[25] A rise in the blood glucose concentration causes them to release insulin into the blood, which has an effect particularly on the liver, but also on fat and muscle cells, causing these tissues to remove glucose from the blood. When the blood sugar falls the pancreatic beta cells cease insulin production, but, instead, stimulate the neighboring pancreatic alpha cells to release glucagon into the blood.[25] This, in turn, causes the liver to release glucose into the blood by breaking down stored glycogen, and by means of gluconeogenesis. If the fall in the blood glucose level is particularly rapid or severe, other glucose sensors cause the release of epinephrine from the adrenal glands into the blood. This has the same action as glucagon on glucose metabolism, but its effect is more pronounced.[25] In the liver glucagon and epinephrine cause the phosphorylation of the key, rate limiting enzymes of glycolysis, fatty acid synthesis, cholesterol synthesis, gluconeogenesis, and glycogenolysis. Insulin has the opposite effect on these enzymes.[26] The phosphorylation and dephosphorylation of these enzymes (ultimately in response to the glucose level in the blood) is the dominant manner by which these pathways are controlled in the liver, fat, and muscle cells. Thus the phosphorylation of phosphofructokinase inhibits glycolysis, whereas its dephosphorylation through the action of insulin stimulates glycolysis.[26]
In addition hexokinase and glucokinase act independently of the hormonal effects as controls at the entry points of glucose into the cells of different tissues. Hexokinase responds to the glucose-6-phosphate (G6P) level in the cell, or, in the case of glucokinase, to the blood sugar level in the blood to impart entirely intracellular controls of the glycolytic pathway in different tissues (see below).[26]
When glucose has been converted into G6P by hexokinase or glucokinase, it can either be converted to glucose-1-phosphate (G1P) for conversion to glycogen, or it is alternatively converted by glycolysis to pyruvate, which enters the mitochondrion where it is converted into acetyl-CoA and then into citrate. Excess citrate is exported from the mitochondrion back into the cytosol, where ATP citrate lyase regenerates acetyl-CoA and oxaloacetate (OAA). The acetyl-CoA is then used for fatty acid synthesis and cholesterol synthesis, two important ways of utilizing excess glucose when its concentration is high in blood. The rate limiting enzymes catalyzing these reactions perform these functions when they have been dephosphorylated through the action of insulin on the liver cells. Between meals, during fasting, exercise or hypoglycemia, glucagon and epinephrine are released into the blood. This causes liver glycogen to be converted back to G6P, and then converted to glucose by the liver-specific enzyme glucose 6-phosphatase and released into the blood. Glucagon and epinephrine also stimulate gluconeogenesis, which coverts non-carbohydrate substrates into G6P, which joins the G6P derived from glycogen, or substitutes for it when the liver glycogen store have been depleted. This is critical for brain function, since the brain utilizes glucose as an energy source under most conditions.[27] The simultaneously phosphorylation of, particularly, phosphofructokinase, but also, to a certain extent pyruvate kinase, prevents glycolysis occurring at the same time as gluconeogenesis and glycogenolysis.
Hexokinase and glucokinase
All cells contain the enzyme hexokinase, which catalyzes the conversion of glucose that has entered the cell into glucose-6-phosphate (G6P). Since the cell membrane is impervious to G6P, hexokinase essentially acts to transport glucose into the cells from which it can then no longer escape. Hexokinase is inhibited by high levels of G6P in the cell. Thus the rate of entry of glucose into cells partially depends on how fast G6P can be disposed of by glycolysis, and by glycogen synthesis (in the cells which store glycogen, namely liver and muscles).[26][28]
Glucokinase, unlike hexokinase, is not inhibited by G6P. It occurs in liver cells, and will only phosphorylate the glucose entering the cell to form glucose-6-phosphate (G6P), when the sugar in the blood is abundant. This being the first step in the glycolytic pathway in the liver, it therefore imparts an additional layer of control of the glycolytic pathway in this organ.[26]
Phosphofructokinase
Phosphofructokinase is an important control point in the glycolytic pathway, since it is one of the irreversible steps and has key allosteric effectors, AMP and fructose 2,6-bisphosphate (F2,6BP).
Fructose 2,6-bisphosphate (F2,6BP) is a very potent activator of phosphofructokinase (PFK-1) that is synthesized when F6P is phosphorylated by a second phosphofructokinase (PFK2). In the liver, when blood sugar is low and glucagon elevates cAMP, PFK2 is phosphorylated by protein kinase A. The phosphorylation inactivates PFK2, and another domain on this protein becomes active as fructose bisphosphatase-2, which converts F2,6BP back to F6P. Both glucagon and epinephrine cause high levels of cAMP in the liver. The result of lower levels of liver fructose-2,6-bisphosphate is a decrease in activity of phosphofructokinase and an increase in activity of fructose 1,6-bisphosphatase, so that gluconeogenesis (in essence, "glycolysis in reverse") is favored. This is consistent with the role of the liver in such situations, since the response of the liver to these hormones is to release glucose to the blood.
ATP competes with AMP for the allosteric effector site on the PFK enzyme. ATP concentrations in cells are much higher than those of AMP, typically 100-fold higher,[29] but the concentration of ATP does not change more than about 10% under physiological conditions, whereas a 10% drop in ATP results in a 6-fold increase in AMP.[30] Thus, the relevance of ATP as an allosteric effector is questionable. An increase in AMP is a consequence of a decrease in energy charge in the cell.
Citrate inhibits phosphofructokinase when tested in vitro by enhancing the inhibitory effect of ATP. However, it is doubtful that this is a meaningful effect in vivo, because citrate in the cytosol is utilized mainly for conversion to acetyl-CoA for fatty acid and cholesterol synthesis.
TIGAR, a p53 induced enzyme, is responsible for the regulation of Phosphofructokinase and acts to protect against oxidative stress.[31] TIGAR is a single enzyme with dual function that regulates F2,6BP. It can behave as a phosphatase (Fructuose-2,6-Bisphosphatase) which cleaves the phosphate at carbon-2 producing F6P. It can also behave as a kinase (PFK2) adding a phosphate onto carbon-2 of F6P which produces F2,6BP. In humans, the TIGAR protein is encoded by C12orf5 gene. The TIGAR enzyme will hinder the forward progression of glycolysis, by creating a build up of fructose-6-phosphate (F6P) which is isomerized into glucose-6-phosphate (G6P). The accumulation of G6P will shunt carbons into the pentose phosphate pathway.[32][33]
Pyruvate kinase
Pyruvate kinase enzyme catalyzes the last step of glycolysis, in which pyruvate and ATP are formed. Pyruvate kinase catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to ADP, yielding one molecule of pyruvate and one molecule of ATP.
Liver pyruvate kinase is indirectly regulated by epinephrine and glucagon, through protein kinase A. This protein kinase phosphorylates liver pyruvate kinase to deactivate it. Muscle pyruvate kinase is not inhibited by epinephrine activation of protein kinase A. Glucagon signals fasting (no glucose available). Thus, glycolysis is inhibited in the liver but unaffected in muscle when fasting. An increase in blood sugar leads to secretion of insulin, which activates phosphoprotein phosphatase I, leading to dephosphorylation and activation of pyruvate kinase. These controls prevent pyruvate kinase from being active at the same time as the enzymes that catalyze the reverse reaction (pyruvate carboxylase and phosphoenolpyruvate carboxykinase), preventing a futile cycle.
Post-glycolysis processes
The overall process of glycolysis is:
- Glucose + 2 NAD+ + 2 ADP + 2 Pi → 2 Pyruvate + 2 NADH + 2 H+ + 2 ATP
If glycolysis were to continue indefinitely, all of the NAD+ would be used up, and glycolysis would stop. To allow glycolysis to continue, organisms must be able to oxidize NADH back to NAD+. How this is performed depends on which external electron acceptor is available.
Anoxic regeneration of NAD+
One method of doing this is to simply have the pyruvate do the oxidation; in this process, pyruvate is converted to lactate (the conjugate base of lactic acid) in a process called lactic acid fermentation:
- Pyruvate + NADH + H+ → Lactate + NAD+
This process occurs in the bacteria involved in making yogurt (the lactic acid causes the milk to curdle). This process also occurs in animals under hypoxic (or partially anaerobic) conditions, found, for example, in overworked muscles that are starved of oxygen. In many tissues, this is a cellular last resort for energy; most animal tissue cannot tolerate anaerobic conditions for an extended period of time.
Some organisms, such as yeast, convert NADH back to NAD+ in a process called ethanol fermentation. In this process, the pyruvate is converted first to acetaldehyde and carbon dioxide, and then to ethanol.
Lactic acid fermentation and ethanol fermentation can occur in the absence of oxygen. This anaerobic fermentation allows many single-cell organisms to use glycolysis as their only energy source.
Anoxic regeneration of NAD+ is only an effective means of energy production during short, intense exercise in vertebrates, for a period ranging from 10 seconds to 2 minutes during a maximal effort in humans. (At lower exercise intensities it can sustain muscle activity in diving animals, such as seals, whales and other aquatic vertebrates, for very much longer periods of time.) Under these conditions NAD+ is replenished by NADH donating its electrons to pyruvate to form lactate. This produces 2 ATP molecules per glucose molecule, or about 5% of glucose's energy potential (38 ATP molecules in bacteria). But the speed at which ATP is produced in this manner is about 100 times that of oxidative phosphorylation. The pH in the cytoplasm quickly drops when hydrogen ions accumulate in the muscle, eventually inhibiting the enzymes involved in glycolysis.
The burning sensation in muscles during hard exercise can be attributed to the release of hydrogen ions during the shift to glucose fermentation from glucose oxidation to carbon dioxide and water, when aerobic metabolism can no longer keep pace with the energy demands of the muscles. These hydrogen ions form a part of lactic acid. The body falls back on this less efficient but faster method of producing ATP under low oxygen conditions. This is thought to have been the primary means of energy production in earlier organisms before oxygen reached high concentrations in the atmosphere between 2000 and 2500 million years ago (see diagram above right), and thus would represent a more ancient form of energy production than the aerobic replenishment of NAD+ in cells.
The liver in mammals gets rid of this excess lactate by transforming it back into pyruvate under aerobic conditions; see Cori cycle.
Fermentation of pyruvate to lactate is sometimes also called "anaerobic glycolysis", however, glycolysis ends with the production of pyruvate regardless of the presence or absence of oxygen.
In the above two examples of fermentation, NADH is oxidized by transferring two electrons to pyruvate. However, anaerobic bacteria use a wide variety of compounds as the terminal electron acceptors in cellular respiration: nitrogenous compounds, such as nitrates and nitrites; sulfur compounds, such as sulfates, sulfites, sulfur dioxide, and elemental sulfur; carbon dioxide; iron compounds; manganese compounds; cobalt compounds; and uranium compounds.
Aerobic regeneration of NAD+, and disposal of pyruvate
In aerobic organisms, a complex mechanism has been developed to use the oxygen in air as the final electron acceptor.
- Firstly, the NADH + H+ generated by glycolysis has to be transferred to the mitochondrion to be oxidized, and thus to regenerate the NAD+ necessary for glycolysis to continue. However the inner mitochondrial membrane is impermeable to NADH and NAD+.[34] Use is therefore made of two “shuttles” to transport the electrons from NADH across the mitochondrial membrane. They are the malate-aspartate shuttle and the glycerol phosphate shuttle. In the former the electrons from NADH are transferred to cytosolic oxaloacetate to form malate. The malate then traverses the inner mitochondrial membrane into the mitochondrial matrix, where it is reoxidized by NAD+ forming intra-mitochondrial oxaloacetate and NADH. The oxaloacetate is then re-cycled to the cytosol via its conversion to aspartate which is readily transported out of the mitochondrion. In the glycerol phosphate shuttle electrons from cytosolic NADH are transferred to dihydroxyacetone to form glycerol-3-phosphate which readily traverses the outer mitochondrial membrane. Glycerol-3-phosphate is then reoxidized to dihydroxyacetone, donating its electrons to FAD instead of NAD+.[34] This reaction takes place on the inner mitochondrial membrane, allowing FADH2 to donate its electrons directly to coenzyme Q (ubiquinone) which is part of the electron transport chain which ultimately transfers electrons to molecular oxygen (O2), with the formation of water, and the release of energy eventually captured in the form of ATP.
- The glycolytic end-product, pyruvate (plus NAD+) is converted to acetyl-CoA, CO2 and NADH + H+ within the mitochondria in a process called pyruvate decarboxylation.
- The resulting acetyl-CoA enters the citric acid cycle (or Krebs Cycle), where the acetyl group of the acetyl-CoA is converted into carbon dioxide by two decarboxylation reactions with the formation of yet more intra-mitochondrial NADH + H+.
- The intra-mitochondrial NADH + H+ is oxidized to NAD+ by the electron transport chain, using oxygen as the final electron acceptor to form water. The energy released during this process is used to create a hydrogen ion (or proton) gradient across the inner membrane of the mitochondrion.
- Finally, the proton gradient is used to produce about 2.5 ATP for every NADH + H+ oxidized in a process called oxidative phosphorylation.[34]
Conversion of carbohydrates into fatty acids and cholesterol
The pyruvate produced by glycolysis is an important intermediary in the conversion of carbohydrates into fatty acids and cholesterol.[35] This occurs via the conversion of pyruvate into acetyl-CoA in the mitochondrion. However, this acetyl CoA needs to be transported into cytosol where the synthesis of fatty acids and cholesterol occurs. This cannot occur directly. To obtain cytosolic acetyl-CoA, citrate (produced by the condensation of acetyl CoA with oxaloacetate) is removed from the citric acid cycle and carried across the inner mitochondrial membrane into the cytosol.[35] There it is cleaved by ATP citrate lyase into acetyl-CoA and oxaloacetate. The oxaloacetate is returned to mitochondrion as malate (and then back into oxaloacetate to transfer more acetyl-CoA out of the mitochondrion). The cytosolic acetyl-CoA can be carboxylated by acetyl-CoA carboxylase into malonyl CoA, the first committed step in the synthesis of fatty acids, or it can be combined with acetoacetyl-CoA to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) which is the rate limiting step controlling the synthesis of cholesterol.[36] Cholesterol can be used as is, as a structural component of cellular membranes, or it can be used to synthesize the steroid hormones, bile salts, and vitamin D.[28][35][36]
Conversion of pyruvate into oxaloacetate for the citric acid cycle
Pyruvate molecules produced by glycolysis are actively transported across the inner mitochondrial membrane, and into the matrix where they can either be oxidized and combined with coenzyme A to form CO2, acetyl-CoA, and NADH,[28] or they can be carboxylated (by pyruvate carboxylase) to form oxaloacetate. This latter reaction "fills up" the amount of oxaloacetate in the citric acid cycle, and is therefore an anaplerotic reaction (from the Greek meaning to "fill up"), increasing the cycle’s capacity to metabolize acetyl-CoA when the tissue's energy needs (e.g. in heart and skeletal muscle) are suddenly increased by activity.[37] In the citric acid cycle all the intermediates (e.g. citrate, iso-citrate, alpha-ketoglutarate, succinate, fumarate, malate and oxaloacetate) are regenerated during each turn of the cycle. Adding more of any of these intermediates to the mitochondrion therefore means that that additional amount is retained within the cycle, increasing all the other intermediates as one is converted into the other. Hence the addition of oxaloacetate greatly increases the amounts of all the citric acid intermediates, thereby increasing the cycle's capacity to metabolize acetyl CoA, converting its acetate component into CO2 and water, with the release of enough energy to form 11 ATP and 1 GTP molecule for each additional molecule of acetyl CoA that combines with oxaloacetate in the cycle.[37]
To cataplerotically remove oxaloacetate from the citric cycle, malate can be transported from the mitochondrion into the cytoplasm, decreasing the amount of oxaloacetate that can be regenerated.[37] Furthermore, citric acid intermediates are constantly used to form a variety of substances such as the purines, pyrimidines and porphyrins.[37]
Intermediates for other pathways
This article concentrates on the catabolic role of glycolysis with regard to converting potential chemical energy to usable chemical energy during the oxidation of glucose to pyruvate. Many of the metabolites in the glycolytic pathway are also used by anabolic pathways, and, as a consequence, flux through the pathway is critical to maintain a supply of carbon skeletons for biosynthesis.
The following metabolic pathways are all strongly reliant on glycolysis as a source of metabolites: and many more.
- Pentose phosphate pathway, which begins with the dehydrogenation of glucose-6-phosphate, the first intermediate to be produced by glycolysis, produces various pentose sugars, and NADPH for the synthesis of fatty acids and cholesterol.
- Glycogen synthesis also starts with glucose-6-phosphate at the beginning of the glycolytic pathway.
- Glycerol, for the formation of triglycerides and phospholipids, is produced from the glycolytic intermediate glyceraldehyde-3-phosphate.
- Various post-glycolytic pathways:
- Fatty acid synthesis
- Cholesterol synthesis
- The citric acid cycle which in turn leads to:
- Amino acid synthesis
- Nucleotide synthesis
- Tetrapyrrole synthesis
Although gluconeogenesis and glycolysis share many intermediates the one is not functionally a branch or tributary of the other. There are two regulatory steps in both pathways which, when active in the one pathway, are automatically inactive in the other. The two processes can therefore not be simultaneously active.[38] Indeed, if both sets of reactions were highly active at the same time the net result would be the hydrolysis of four high energy phosphate bonds (two ATP and two GTP) per reaction cycle.[38]
NAD+ is the oxidizing agent in glycolysis, as it is in most other energy yielding metabolic reactions (e.g. beta-oxidation of fatty acids, and during the citric acid cycle). The NADH thus produced is primarily used to ultimately transfer electrons to O2 to produce water, or, when O2 is not available, to produced compounds such as lactate or ethanol (see Anoxic regeneration of NAD+ above). NADH is rarely used for synthetic processes, the notable exception being gluconeogenesis. During fatty acid and cholesterol synthesis the reducing agent is NADPH. This difference exemplifies a general principle that NADPH is consumed during biosynthetic reactions, whereas NADH is generated in energy-yielding reactions.[38] The source of the NADPH is two-fold. When malate is oxidatively decarboxylated by “NADP+-linked malic enzyme" pyruvate, CO2 and NADPH are formed. NADPH is also formed by the pentose phosphate pathway which converts glucose into ribose, which can be used in synthesis of nucleotides and nucleic acids, or it can be catabolized to pyruvate.[38]
Glycolysis in disease
Diabetes
Cellular uptake of glucose occurs in response to insulin signals, and glucose is subsequently broken down through glycolysis, lowering blood sugar levels. However, the low insulin levels seen in diabetes result in hyperglycemia, where glucose levels in the blood rise and glucose is not properly taken up by cells. Hepatocytes further contribute to this hyperglycemia through gluconeogenesis. Glycolysis in hepatocytes controls hepatic glucose production, and when glucose is overproduced by the liver without having a means of being broken down by the body, hyperglycemia results.[39]
Genetic diseases
Glycolytic mutations are generally rare due to importance of the metabolic pathway, this means that the majority of occurring mutations result in an inability for the cell to respire, and therefore cause the death of the cell at an early stage. However, some mutations are seen with one notable example being Pyruvate kinase deficiency, leading to chronic hemolytic anemia.
Cancer
Malignant tumor cells perform glycolysis at a rate that is ten times faster than their noncancerous tissue counterparts.[40] During their genesis, limited capillary support often results in hypoxia (decreased O2 supply) within the tumor cells. Thus, these cells rely on anaerobic metabolic processes such as glycolysis for ATP (adenosine triphosphate). Some tumor cells overexpress specific glycolytic enzymes which result in higher rates of glycolysis.[41] Often these enzymes are Isoenzymes, of traditional glycolysis enzymes, that vary in their susceptibility to traditional feedback inhibition. The increase in glycolytic activity ultimately counteracts the effects of hypoxia by generating sufficient ATP from this anaerobic pathway.[42] This phenomenon was first described in 1930 by Otto Warburg and is referred to as the Warburg effect. The Warburg hypothesis claims that cancer is primarily caused by dysfunctionality in mitochondrial metabolism, rather than because of the uncontrolled growth of cells. A number of theories have been advanced to explain the Warburg effect. One such theory suggests that the increased glycolysis is a normal protective process of the body and that malignant change could be primarily caused by energy metabolism.[43]
This high glycolysis rate has important medical applications, as high aerobic glycolysis by malignant tumors is utilized clinically to diagnose and monitor treatment responses of cancers by imaging uptake of 2-18F-2-deoxyglucose (FDG) (a radioactive modified hexokinase substrate) with positron emission tomography (PET).[44][45]
There is ongoing research to affect mitochondrial metabolism and treat cancer by reducing glycolysis and thus starving cancerous cells in various new ways, including a ketogenic diet.[46][47][48]
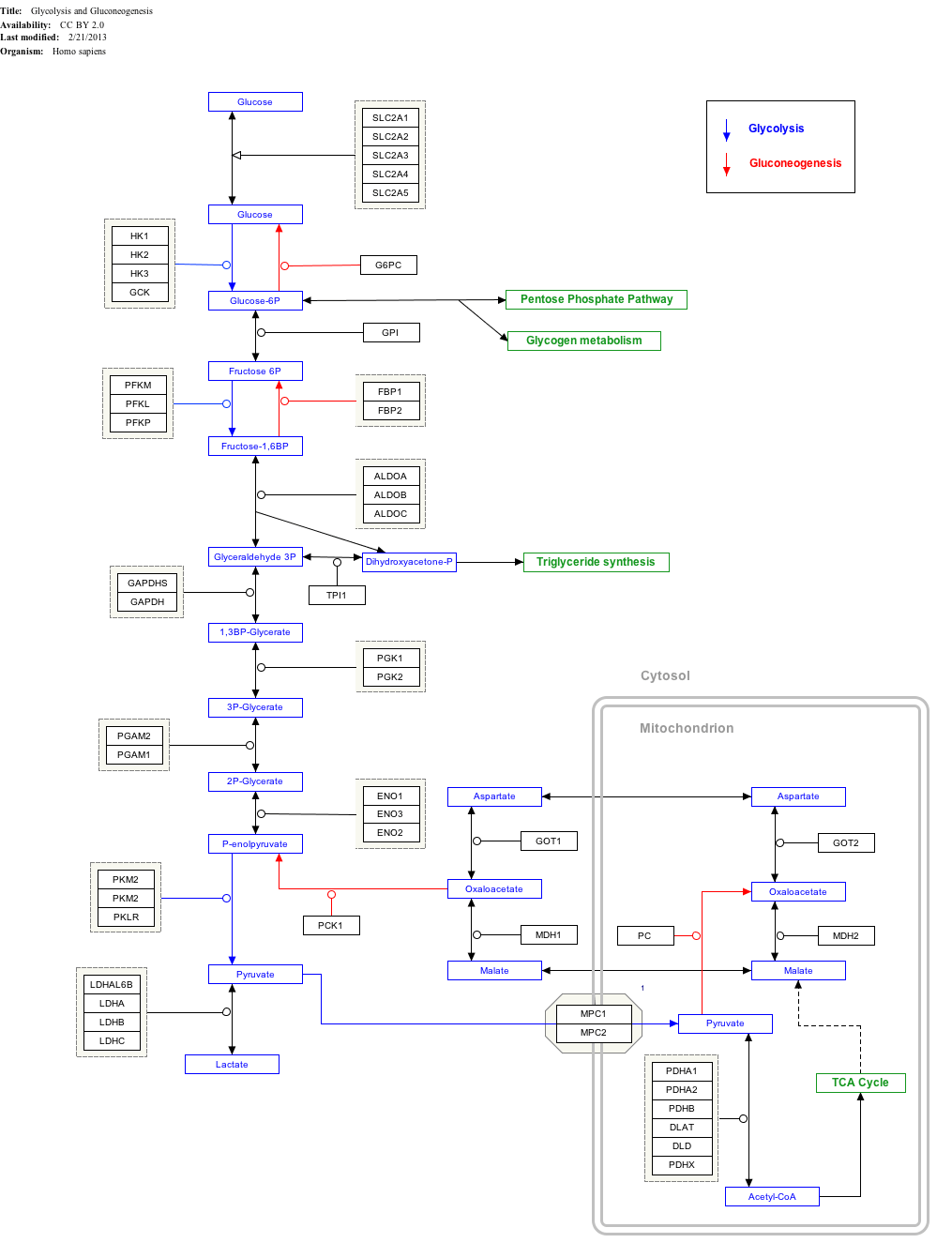
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
Alternative nomenclature
Some of the metabolites in glycolysis have alternative names and nomenclature. In part, this is because some of them are common to other pathways, such as the Calvin cycle.
| This article | Alternative names | Alternative nomenclature | ||
|---|---|---|---|---|
| 1 | Glucose | Glc | Dextrose | |
| 2 | Glucose-6-phosphate | G6P | ||
| 3 | Fructose-6-phosphate | F6P | ||
| 4 | Fructose-1,6-bisphosphate | F1,6BP | Fructose 1,6-diphosphate | FBP, FDP, F1,6DP |
| 5 | Dihydroxyacetone phosphate | DHAP | Glycerone phosphate | |
| 6 | Glyceraldehyde-3-phosphate | GADP | 3-Phosphoglyceraldehyde | PGAL, G3P, GALP,GAP,TP |
| 7 | 1,3-Bisphosphoglycerate | 1,3BPG | Glycerate-1,3-bisphosphate, glycerate-1,3-diphosphate, 1,3-diphosphoglycerate |
PGAP, BPG, DPG |
| 8 | 3-Phosphoglycerate | 3PG | Glycerate-3-phosphate | PGA, GP |
| 9 | 2-Phosphoglycerate | 2PG | Glycerate-2-phosphate | |
| 10 | Phosphoenolpyruvate | PEP | ||
| 11 | Pyruvate | Pyr | Pyruvic acid | |
See also
| Wikimedia Commons has media related to Glycolysis. |
- Carbohydrate catabolism
- Citric acid cycle
- Cori cycle
- Fermentation (biochemistry)
- Gluconeogenesis
- Glycolytic oscillation
- Pentose phosphate pathway
- Pyruvate decarboxylation
- Triose kinase
References
- Webster's New International Dictionary of the English Language, 2nd ed. (1937) Merriam Company, Springfield, Mass.
- Glycolysis – Animation and Notes
- Bailey, Regina. "10 Steps of Glycolysis".
- Romano, AH; Conway, T (1996). "Evolution of carbohydrate metabolic pathways". Res Microbiol. 147 (6–7): 448–55. doi:10.1016/0923-2508(96)83998-2. PMID 9084754.
- Keller; Ralser; Turchyn (Apr 2014). "Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean". Mol Syst Biol. 10 (4): 725. doi:10.1002/msb.20145228. PMC 4023395. PMID 24771084.
- Kim BH, Gadd GM. (2011) Bacterial Physiology and Metabolism, 3rd edition.
- Lane, A. N.; Fan, T. W. -M.; Higashi, R. M. (2009). "Metabolic acidosis and the importance of balanced equations". Metabolomics. 5 (2): 163–165. doi:10.1007/s11306-008-0142-2.
- Barnett JA (April 2003). "A history of research on yeasts 5: the fermentation pathway". Yeast. 20 (6): 509–543. doi:10.1002/yea.986. PMID 12722184.
- "Louis Pasteur and Alcoholic Fermentation". www.pasteurbrewing.com. Retrieved 2016-02-23.
- "Yeast, Fermentation, Beer, Wine". www.nature.com. Retrieved 2016-02-23.
- Kohler, Robert (1971-03-01). "The background to Eduard Buchner's discovery of cell-free fermentation". Journal of the History of Biology. 4 (1): 35–61. doi:10.1007/BF00356976. ISSN 0022-5010. PMID 11609437.
- "Eduard Buchner - Biographical". www.nobelprize.org. Retrieved 2016-02-23.
- Cornish-Bowden, Athel (1997). "Harden and Young's Discovery of Fructose 1,6-Bisphosphate". New Beer in an Old Bottle: Eduard Buchner and the Growth of Biochemical Knowledge. Valencia, Spain. pp. 135–148.
- Palmer, Grahm. "Chapter 3". Bios 302. http://www.bioc.rice.edu/~graham/Bios302/chapters/.
- Cornish-Bowden, Athel (1997). "Harden and Young's Discovery of Fructose 1,6-Bisphosphate". New Beer in an Old Bottle: Eduard Buchner and the Growth of Biochemical Knowledge. Valencia, Spain. pp. 151–158.
- "Otto Meyerhof - Biographical". www.nobelprize.org. Retrieved 2016-02-23.
- Kresge, Nicole; Simoni, Robert D.; Hill, Robert L. (2005-01-28). "Otto Fritz Meyerhof and the Elucidation of the Glycolytic Pathway". Journal of Biological Chemistry. 280 (4): e3. ISSN 0021-9258. PMID 15665335.
- "Embden, Gustav – Dictionary definition of Embden, Gustav | Encyclopedia.com: FREE online dictionary". www.encyclopedia.com. Retrieved 2016-02-23.
- Reeves, R. E.; South D. J.; Blytt H. J.; Warren L. G. (1974). "Pyrophosphate: D-fructose 6-phosphate 1-phosphotransferase. A new enzyme with the glycolytic function 6-phosphate 1-phosphotransferase". J Biol Chem. 249 (24): 7737–7741. PMID 4372217.
- Selig, M.; Xavier K. B.; Santos H.; Schönheit P. (1997). "Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga". Arch Microbiol. 167 (4): 217–232. doi:10.1007/BF03356097. PMID 9075622.
- Garrett, Reginald H.; Grisham, Charles M. (2012). Biochemistry. Cengage Learning; 5 edition. ISBN 978-1-133-10629-6.
- Berg, J. M.; Tymoczko, J. L.; Stryer, L. (2007). Biochemistry (6th ed.). New York: Freeman. p. 622. ISBN 978-0716787242.
- Garrett, R.; Grisham, C. M. (2005). Biochemistry (3rd ed.). Belmont, CA: Thomson Brooks/Cole. p. 584. ISBN 978-0-534-49033-1.
- Garrett, R.; Grisham, C. M. (2005). Biochemistry (3rd ed.). Belmont, CA: Thomson Brooks/Cole. pp. 582–583. ISBN 978-0-534-49033-1.
- Koeslag, Johan H.; Saunders, Peter T.; Terblanche, Elmarie (2003). "Topical Review: A reappraisal of the blood glucose homeostat which comprehensively explains the type 2 diabetes-syndrome X complex". Journal of Physiology. 549 (Pt 2): 333–346. doi:10.1113/jphysiol.2002.037895. PMC 2342944. PMID 12717005.
- Stryer, Lubert (1995). "Glycolysis.". In: Biochemistry (Fourth ed.). New York: W.H. Freeman and Company. pp. 483–508. ISBN 0-7167-2009-4.
- Stryer, Lubert (1995). Biochemistry (Fourth ed.). New York: W.H. Freeman and Company. p. 773. ISBN 0-7167-2009-4.
- Voet, Donald; Judith G. Voet; Charlotte W. Pratt (2006). Fundamentals of Biochemistry, 2nd Edition. John Wiley and Sons, Inc. pp. 547, 556. ISBN 978-0-471-21495-3.
- Beis, I.; Newsholme, E. A. (1975). "The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates". Biochem J. 152 (1): 23–32. doi:10.1042/bj1520023. PMC 1172435. PMID 1212224.
- Voet D., and Voet J. G. (2004). Biochemistry 3rd Edition (New York, John Wiley & Sons, Inc.).
- Lackie, John (2010). TIGAR. Oxford Reference Online: Oxford University Press. ISBN 9780199549351.
- Bensaad, Karim (July 16, 2006). "TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis". Cell. 126 (I): 107–120. doi:10.1016/j.cell.2006.05.036. PMID 16839880.
- "TIGAR TP53 induced glycolysis regulatory phosphatase [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2018-05-17.
- Stryer, Lubert (1995). "Oxidative phosphorylation.". In: Biochemistry (Fourth ed.). New York: W.H. Freeman and Company. pp. 537–549. ISBN 0-7167-2009-4.
- Stryer, Lubert (1995). "Fatty acid metabolism.". In: Biochemistry (Fourth ed.). New York: W.H. Freeman and Company. pp. 603–628. ISBN 0-7167-2009-4.
- Stryer, Lubert (1995). "Biosynthesis of membrane lipids and steroids.". In: Biochemistry (Fourth ed.). New York: W.H. Freeman and Company. pp. 691–707. ISBN 0-7167-2009-4.
- Stryer, Lubert (1995). "Citric acid cycle.". In: Biochemistry (Fourth ed.). New York: W.H. Freeman and Company. pp. 509–527, 569–579, 614–616, 638–641, 732–735, 739–748, 770–773. ISBN 0-7167-2009-4.
- Stryer, Lubert (1995). Biochemistry (Fourth ed.). New York: W.H. Freeman and Company. pp. 559–565, 574–576, 614–623. ISBN 0-7167-2009-4.
- Guo, Xin; Li, Honggui; Xu, Hang; Woo, Shihlung; Dong, Hui; Lu, Fuer; Lange, Alex J.; Wu, Chaodong (2012-08-01). "Glycolysis in the control of blood glucose homeostasis". Acta Pharmaceutica Sinica B. 2 (4): 358–367. doi:10.1016/j.apsb.2012.06.002. ISSN 2211-3835.
- Alfarouk, KO; Verduzco, D; Rauch, C; Muddathir, AK; Adil, HH; Elhassan, GO; Ibrahim, ME; David Polo Orozco, J; Cardone, RA; Reshkin, SJ; Harguindey, S (2014). "Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question". Oncoscience. 1 (12): 777–802. doi:10.18632/oncoscience.109. PMC 4303887. PMID 25621294.
- Alfarouk, KO; Shayoub, ME; Muddathir, AK; Elhassan, GO; Bashir, AH (22 July 2011). "Evolution of Tumor Metabolism might Reflect Carcinogenesis as a Reverse Evolution process (Dismantling of Multicellularity)". Cancers. 3 (3): 3002–17. doi:10.3390/cancers3033002. PMC 3759183. PMID 24310356.
- Cox, David L. Nelson, Michael M. (2005). Lehninger principles of biochemistry (4th ed.). New York: W.H. Freeman. ISBN 978-0-7167-4339-2.
- Gold, Joseph (October 2011). "What is Cancer?". Retrieved September 8, 2012. Cite journal requires
|journal=(help) - "4320139 549..559" (PDF). Retrieved December 5, 2005.
- "PET Scan: PET Scan Info Reveals ..." Retrieved December 5, 2005.
- Schwartz, L; Seyfried, T; Alfarouk, KO; Da Veiga Moreira, J; Fais, S (April 2017). "Out of Warburg effect: An effective cancer treatment targeting the tumor specific metabolism and dysregulated pH". Seminars in Cancer Biology. 43: 134–138. doi:10.1016/j.semcancer.2017.01.005. PMID 28122260.
- Schwartz, L; Supuran, CT; Alfarouk, KO (2017). "The Warburg Effect and the Hallmarks of Cancer". Anti-Cancer Agents in Medicinal Chemistry. 17 (2): 164–170. doi:10.2174/1871520616666161031143301. PMID 27804847.
- Maroon, J; Bost J; Amos A; Zuccoli G (May 2013). "Restricted Calorie Ketogenic Diet for the Treatment of Glioblastoma Multiforme". Journal of Child Neurology. 28 (8): 1002–1008. doi:10.1177/0883073813488670. PMID 23670248.
External links
- A Detailed Glycolysis Animation provided by IUBMB (Adobe Flash Required)
- The Glycolytic enzymes in Glycolysis at RCSB PDB
- Glycolytic cycle with animations at wdv.com
- Metabolism, Cellular Respiration and Photosynthesis - The Virtual Library of Biochemistry, Molecular Biology and Cell Biology at biochemweb.net
- The chemical logic behind glycolysis at ufp.pt
- Expasy biochemical pathways poster at ExPASy
- MedicalMnemonics.com: 317 5468
- metpath: Interactive representation of glycolysis
| Library resources about Glycolysis |

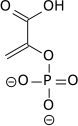
.svg.png)