Fungus
A fungus (plural: fungi[3] or funguses[4]) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, fungi, which is separate from the other eukaryotic life kingdoms of plants and animals.
| Fungi | |
|---|---|
 | |
Clockwise from top left:
| |
| Scientific classification | |
| (unranked): | Opisthokonta |
| (unranked): | Holomycota |
| (unranked): | Zoosporia |
| Kingdom: | Fungi (L.) R.T.Moore[1] |
| Subkingdoms/Phyla/Subphyla[2] | |
Dikarya (inc. Deuteromycota)
Subphyla incertae sedis
| |
A characteristic that places fungi in a different kingdom from plants, bacteria, and some protists is chitin in their cell walls. Similar to animals, fungi are heterotrophs; they acquire their food by absorbing dissolved molecules, typically by secreting digestive enzymes into their environment. Fungi do not photosynthesize. Growth is their means of mobility, except for spores (a few of which are flagellated), which may travel through the air or water. Fungi are the principal decomposers in ecological systems. These and other differences place fungi in a single group of related organisms, named the Eumycota (true fungi or Eumycetes), which share a common ancestor (form a monophyletic group), an interpretation that is also strongly supported by molecular phylogenetics. This fungal group is distinct from the structurally similar myxomycetes (slime molds) and oomycetes (water molds). The discipline of biology devoted to the study of fungi is known as mycology (from the Greek μύκης mykes, mushroom). In the past, mycology was regarded as a branch of botany, although it is now known fungi are genetically more closely related to animals than to plants.
Abundant worldwide, most fungi are inconspicuous because of the small size of their structures, and their cryptic lifestyles in soil or on dead matter. Fungi include symbionts of plants, animals, or other fungi and also parasites. They may become noticeable when fruiting, either as mushrooms or as molds. Fungi perform an essential role in the decomposition of organic matter and have fundamental roles in nutrient cycling and exchange in the environment. They have long been used as a direct source of human food, in the form of mushrooms and truffles; as a leavening agent for bread; and in the fermentation of various food products, such as wine, beer, and soy sauce. Since the 1940s, fungi have been used for the production of antibiotics, and, more recently, various enzymes produced by fungi are used industrially and in detergents. Fungi are also used as biological pesticides to control weeds, plant diseases and insect pests. Many species produce bioactive compounds called mycotoxins, such as alkaloids and polyketides, that are toxic to animals including humans. The fruiting structures of a few species contain psychotropic compounds and are consumed recreationally or in traditional spiritual ceremonies. Fungi can break down manufactured materials and buildings, and become significant pathogens of humans and other animals. Losses of crops due to fungal diseases (e.g., rice blast disease) or food spoilage can have a large impact on human food supplies and local economies.
The fungus kingdom encompasses an enormous diversity of taxa with varied ecologies, life cycle strategies, and morphologies ranging from unicellular aquatic chytrids to large mushrooms. However, little is known of the true biodiversity of Kingdom Fungi, which has been estimated at 2.2 million to 3.8 million species.[5] Of these, only about 120,000 have been described, with over 8,000 species known to be detrimental to plants and at least 300 that can be pathogenic to humans.[6] Ever since the pioneering 18th and 19th century taxonomical works of Carl Linnaeus, Christian Hendrik Persoon, and Elias Magnus Fries, fungi have been classified according to their morphology (e.g., characteristics such as spore color or microscopic features) or physiology. Advances in molecular genetics have opened the way for DNA analysis to be incorporated into taxonomy, which has sometimes challenged the historical groupings based on morphology and other traits. Phylogenetic studies published in the last decade have helped reshape the classification within Kingdom Fungi, which is divided into one subkingdom, seven phyla, and ten subphyla.
Etymology
The English word fungus is directly adopted from the Latin fungus (mushroom), used in the writings of Horace and Pliny.[7] This in turn is derived from the Greek word sphongos (σφόγγος "sponge"), which refers to the macroscopic structures and morphology of mushrooms and molds;[8] the root is also used in other languages, such as the German Schwamm ("sponge") and Schimmel ("mold").[9]
The word mycology is derived from the Greek mykes (μύκης "mushroom") and logos (λόγος "discourse").[10] It denotes the scientific study of fungi. The Latin adjectival form of "mycology" (mycologicæ) appeared as early as 1796 in a book on the subject by Christiaan Hendrik Persoon.[11] The word appeared in English as early as 1824 in a book by Robert Kaye Greville.[12] In 1836 the English naturalist Miles Joseph Berkeley's publication The English Flora of Sir James Edward Smith, Vol. 5. also refers to mycology as the study of fungi.[8][13]
A group of all the fungi present in a particular area or geographic region is known as mycobiota (plural noun, no singular), e.g., "the mycobiota of Ireland".[14]
Characteristics
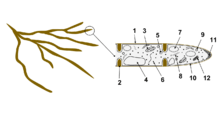
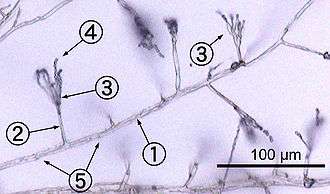
- Hyphal wall
- Septum
- Mitochondrion
- Vacuole
- Ergosterol crystal
- Ribosome
- Nucleus
- Endoplasmic reticulum
- Lipid body
- Plasma membrane
- Spitzenkörper
- Golgi apparatus
Before the introduction of molecular methods for phylogenetic analysis, taxonomists considered fungi to be members of the plant kingdom because of similarities in lifestyle: both fungi and plants are mainly immobile, and have similarities in general morphology and growth habitat. Like plants, fungi often grow in soil and, in the case of mushrooms, form conspicuous fruit bodies, which sometimes resemble plants such as mosses. The fungi are now considered a separate kingdom, distinct from both plants and animals, from which they appear to have diverged around one billion years ago (around the start of the Neoproterozoic Era).[15][16] Some morphological, biochemical, and genetic features are shared with other organisms, while others are unique to the fungi, clearly separating them from the other kingdoms:
Shared features:
- With other eukaryotes: Fungal cells contain membrane-bound nuclei with chromosomes that contain DNA with noncoding regions called introns and coding regions called exons. Fungi have membrane-bound cytoplasmic organelles such as mitochondria, sterol-containing membranes, and ribosomes of the 80S type.[17] They have a characteristic range of soluble carbohydrates and storage compounds, including sugar alcohols (e.g., mannitol), disaccharides, (e.g., trehalose), and polysaccharides (e.g., glycogen, which is also found in animals[18]).
- With animals: Fungi lack chloroplasts and are heterotrophic organisms and so require preformed organic compounds as energy sources.[19]
- With plants: Fungi have a cell wall[20] and vacuoles.[21] They reproduce by both sexual and asexual means, and like basal plant groups (such as ferns and mosses) produce spores. Similar to mosses and algae, fungi typically have haploid nuclei.[22]
- With euglenoids and bacteria: Higher fungi, euglenoids, and some bacteria produce the amino acid L-lysine in specific biosynthesis steps, called the α-aminoadipate pathway.[23][24]
- The cells of most fungi grow as tubular, elongated, and thread-like (filamentous) structures called hyphae, which may contain multiple nuclei and extend by growing at their tips. Each tip contains a set of aggregated vesicles—cellular structures consisting of proteins, lipids, and other organic molecules—called the Spitzenkörper.[25] Both fungi and oomycetes grow as filamentous hyphal cells.[26] In contrast, similar-looking organisms, such as filamentous green algae, grow by repeated cell division within a chain of cells.[18] There are also single-celled fungi (yeasts) that do not form hyphae, and some fungi have both hyphal and yeast forms.[27]
- In common with some plant and animal species, more than 70 fungal species display bioluminescence.[28]
Unique features:
- Some species grow as unicellular yeasts that reproduce by budding or fission. Dimorphic fungi can switch between a yeast phase and a hyphal phase in response to environmental conditions.[27]
- The fungal cell wall is composed of glucans and chitin; while glucans are also found in plants and chitin in the exoskeleton of arthropods,[29][30] fungi are the only organisms that combine these two structural molecules in their cell wall. Unlike those of plants and oomycetes, fungal cell walls do not contain cellulose.[31]

Most fungi lack an efficient system for the long-distance transport of water and nutrients, such as the xylem and phloem in many plants. To overcome this limitation, some fungi, such as Armillaria, form rhizomorphs,[32] which resemble and perform functions similar to the roots of plants. As eukaryotes, fungi possess a biosynthetic pathway for producing terpenes that uses mevalonic acid and pyrophosphate as chemical building blocks.[33] Plants and some other organisms have an additional terpene biosynthesis pathway in their chloroplasts, a structure fungi and animals do not have.[34] Fungi produce several secondary metabolites that are similar or identical in structure to those made by plants.[33] Many of the plant and fungal enzymes that make these compounds differ from each other in sequence and other characteristics, which indicates separate origins and convergent evolution of these enzymes in the fungi and plants.[33][35]
Diversity
Fungi have a worldwide distribution, and grow in a wide range of habitats, including extreme environments such as deserts or areas with high salt concentrations[36] or ionizing radiation,[37] as well as in deep sea sediments.[38] Some can survive the intense UV and cosmic radiation encountered during space travel.[39] Most grow in terrestrial environments, though several species live partly or solely in aquatic habitats, such as the chytrid fungus Batrachochytrium dendrobatidis, a parasite that has been responsible for a worldwide decline in amphibian populations. This organism spends part of its life cycle as a motile zoospore, enabling it to propel itself through water and enter its amphibian host.[40] Other examples of aquatic fungi include those living in hydrothermal areas of the ocean.[41]
Around 120,000 species of fungi have been described by taxonomists,[42] but the global biodiversity of the fungus kingdom is not fully understood.[42] A 2017 estimate suggests there may be between 2.2 and 3.8 million species.[5] In mycology, species have historically been distinguished by a variety of methods and concepts. Classification based on morphological characteristics, such as the size and shape of spores or fruiting structures, has traditionally dominated fungal taxonomy.[43] Species may also be distinguished by their biochemical and physiological characteristics, such as their ability to metabolize certain biochemicals, or their reaction to chemical tests. The biological species concept discriminates species based on their ability to mate. The application of molecular tools, such as DNA sequencing and phylogenetic analysis, to study diversity has greatly enhanced the resolution and added robustness to estimates of genetic diversity within various taxonomic groups.[44]
Mycology
Mycology is the branch of biology concerned with the systematic study of fungi, including their genetic and biochemical properties, their taxonomy, and their use to humans as a source of medicine, food, and psychotropic substances consumed for religious purposes, as well as their dangers, such as poisoning or infection. The field of phytopathology, the study of plant diseases, is closely related because many plant pathogens are fungi.[45]

The use of fungi by humans dates back to prehistory; Ötzi the Iceman, a well-preserved mummy of a 5,300-year-old Neolithic man found frozen in the Austrian Alps, carried two species of polypore mushrooms that may have been used as tinder (Fomes fomentarius), or for medicinal purposes (Piptoporus betulinus).[46] Ancient peoples have used fungi as food sources–often unknowingly–for millennia, in the preparation of leavened bread and fermented juices. Some of the oldest written records contain references to the destruction of crops that were probably caused by pathogenic fungi.[47]
History
Mycology is a relatively new science that became systematic after the development of the microscope in the 17th century. Although fungal spores were first observed by Giambattista della Porta in 1588, the seminal work in the development of mycology is considered to be the publication of Pier Antonio Micheli's 1729 work Nova plantarum genera.[48] Micheli not only observed spores but also showed that, under the proper conditions, they could be induced into growing into the same species of fungi from which they originated.[49] Extending the use of the binomial system of nomenclature introduced by Carl Linnaeus in his Species plantarum (1753), the Dutch Christian Hendrik Persoon (1761–1836) established the first classification of mushrooms with such skill as to be considered a founder of modern mycology. Later, Elias Magnus Fries (1794–1878) further elaborated the classification of fungi, using spore color and microscopic characteristics, methods still used by taxonomists today. Other notable early contributors to mycology in the 17th–19th and early 20th centuries include Miles Joseph Berkeley, August Carl Joseph Corda, Anton de Bary, the brothers Louis René and Charles Tulasne, Arthur H. R. Buller, Curtis G. Lloyd, and Pier Andrea Saccardo. The 20th century has seen a modernization of mycology that has come from advances in biochemistry, genetics, molecular biology, and biotechnology. The use of DNA sequencing technologies and phylogenetic analysis has provided new insights into fungal relationships and biodiversity, and has challenged traditional morphology-based groupings in fungal taxonomy.[50]
Morphology
Microscopic structures
Most fungi grow as hyphae, which are cylindrical, thread-like structures 2–10 µm in diameter and up to several centimeters in length. Hyphae grow at their tips (apices); new hyphae are typically formed by emergence of new tips along existing hyphae by a process called branching, or occasionally growing hyphal tips fork, giving rise to two parallel-growing hyphae.[51] Hyphae also sometimes fuse when they come into contact, a process called hyphal fusion (or anastomosis). These growth processes lead to the development of a mycelium, an interconnected network of hyphae.[27] Hyphae can be either septate or coenocytic. Septate hyphae are divided into compartments separated by cross walls (internal cell walls, called septa, that are formed at right angles to the cell wall giving the hypha its shape), with each compartment containing one or more nuclei; coenocytic hyphae are not compartmentalized.[52] Septa have pores that allow cytoplasm, organelles, and sometimes nuclei to pass through; an example is the dolipore septum in fungi of the phylum Basidiomycota.[53] Coenocytic hyphae are in essence multinucleate supercells.[54]
Many species have developed specialized hyphal structures for nutrient uptake from living hosts; examples include haustoria in plant-parasitic species of most fungal phyla, and arbuscules of several mycorrhizal fungi, which penetrate into the host cells to consume nutrients.[55]
Although fungi are opisthokonts—a grouping of evolutionarily related organisms broadly characterized by a single posterior flagellum—all phyla except for the chytrids have lost their posterior flagella.[56] Fungi are unusual among the eukaryotes in having a cell wall that, in addition to glucans (e.g., β-1,3-glucan) and other typical components, also contains the biopolymer chitin.[57]
Macroscopic structures

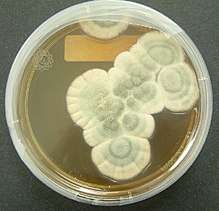
Fungal mycelia can become visible to the naked eye, for example, on various surfaces and substrates, such as damp walls and spoiled food, where they are commonly called molds. Mycelia grown on solid agar media in laboratory petri dishes are usually referred to as colonies. These colonies can exhibit growth shapes and colors (due to spores or pigmentation) that can be used as diagnostic features in the identification of species or groups.[58] Some individual fungal colonies can reach extraordinary dimensions and ages as in the case of a clonal colony of Armillaria solidipes, which extends over an area of more than 900 ha (3.5 square miles), with an estimated age of nearly 9,000 years.[59]
The apothecium—a specialized structure important in sexual reproduction in the ascomycetes—is a cup-shaped fruit body that is often macroscopic and holds the hymenium, a layer of tissue containing the spore-bearing cells.[60] The fruit bodies of the basidiomycetes (basidiocarps) and some ascomycetes can sometimes grow very large, and many are well known as mushrooms.
Growth and physiology

The growth of fungi as hyphae on or in solid substrates or as single cells in aquatic environments is adapted for the efficient extraction of nutrients, because these growth forms have high surface area to volume ratios.[61] Hyphae are specifically adapted for growth on solid surfaces, and to invade substrates and tissues.[62] They can exert large penetrative mechanical forces; for example, many plant pathogens, including Magnaporthe grisea, form a structure called an appressorium that evolved to puncture plant tissues.[63] The pressure generated by the appressorium, directed against the plant epidermis, can exceed 8 megapascals (1,200 psi).[63] The filamentous fungus Paecilomyces lilacinus uses a similar structure to penetrate the eggs of nematodes.[64]
The mechanical pressure exerted by the appressorium is generated from physiological processes that increase intracellular turgor by producing osmolytes such as glycerol.[65] Adaptations such as these are complemented by hydrolytic enzymes secreted into the environment to digest large organic molecules—such as polysaccharides, proteins, and lipids—into smaller molecules that may then be absorbed as nutrients.[66][67][68] The vast majority of filamentous fungi grow in a polar fashion (extending in one direction) by elongation at the tip (apex) of the hypha.[69] Other forms of fungal growth include intercalary extension (longitudinal expansion of hyphal compartments that are below the apex) as in the case of some endophytic fungi,[70] or growth by volume expansion during the development of mushroom stipes and other large organs.[71] Growth of fungi as multicellular structures consisting of somatic and reproductive cells—a feature independently evolved in animals and plants[72]—has several functions, including the development of fruit bodies for dissemination of sexual spores (see above) and biofilms for substrate colonization and intercellular communication.[73]
The fungi are traditionally considered heterotrophs, organisms that rely solely on carbon fixed by other organisms for metabolism. Fungi have evolved a high degree of metabolic versatility that allows them to use a diverse range of organic substrates for growth, including simple compounds such as nitrate, ammonia, acetate, or ethanol.[74][75] In some species the pigment melanin may play a role in extracting energy from ionizing radiation, such as gamma radiation. This form of "radiotrophic" growth has been described for only a few species, the effects on growth rates are small, and the underlying biophysical and biochemical processes are not well known.[37] This process might bear similarity to CO2 fixation via visible light, but instead uses ionizing radiation as a source of energy.[76]
Reproduction

Fungal reproduction is complex, reflecting the differences in lifestyles and genetic makeup within this diverse kingdom of organisms.[77] It is estimated that a third of all fungi reproduce using more than one method of propagation; for example, reproduction may occur in two well-differentiated stages within the life cycle of a species, the teleomorph and the anamorph.[78] Environmental conditions trigger genetically determined developmental states that lead to the creation of specialized structures for sexual or asexual reproduction. These structures aid reproduction by efficiently dispersing spores or spore-containing propagules.
Asexual reproduction
Asexual reproduction occurs via vegetative spores (conidia) or through mycelial fragmentation. Mycelial fragmentation occurs when a fungal mycelium separates into pieces, and each component grows into a separate mycelium. Mycelial fragmentation and vegetative spores maintain clonal populations adapted to a specific niche, and allow more rapid dispersal than sexual reproduction.[79] The "Fungi imperfecti" (fungi lacking the perfect or sexual stage) or Deuteromycota comprise all the species that lack an observable sexual cycle.[80] Deuteromycota is not an accepted taxonomic clade, and is now taken to mean simply fungi that lack a known sexual stage.
Sexual reproduction
Sexual reproduction with meiosis has been directly observed in all fungal phyla except Glomeromycota [81] (genetic analysis suggests meiosis in Glomeromycota as well). It differs in many aspects from sexual reproduction in animals or plants. Differences also exist between fungal groups and can be used to discriminate species by morphological differences in sexual structures and reproductive strategies.[82][83] Mating experiments between fungal isolates may identify species on the basis of biological species concepts.[83] The major fungal groupings have initially been delineated based on the morphology of their sexual structures and spores; for example, the spore-containing structures, asci and basidia, can be used in the identification of ascomycetes and basidiomycetes, respectively. Fungi employ two mating systems: heterothallic species allow mating only between individuals of opposite mating type, whereas homothallic species can mate, and sexually reproduce, with any other individual or itself.[84]
Most fungi have both a haploid and a diploid stage in their life cycles. In sexually reproducing fungi, compatible individuals may combine by fusing their hyphae together into an interconnected network; this process, anastomosis, is required for the initiation of the sexual cycle. Many ascomycetes and basidiomycetes go through a dikaryotic stage, in which the nuclei inherited from the two parents do not combine immediately after cell fusion, but remain separate in the hyphal cells (see heterokaryosis).[85]
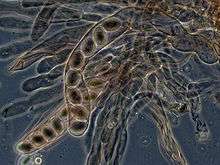
In ascomycetes, dikaryotic hyphae of the hymenium (the spore-bearing tissue layer) form a characteristic hook at the hyphal septum. During cell division, formation of the hook ensures proper distribution of the newly divided nuclei into the apical and basal hyphal compartments. An ascus (plural asci) is then formed, in which karyogamy (nuclear fusion) occurs. Asci are embedded in an ascocarp, or fruiting body. Karyogamy in the asci is followed immediately by meiosis and the production of ascospores. After dispersal, the ascospores may germinate and form a new haploid mycelium.[86]
Sexual reproduction in basidiomycetes is similar to that of the ascomycetes. Compatible haploid hyphae fuse to produce a dikaryotic mycelium. However, the dikaryotic phase is more extensive in the basidiomycetes, often also present in the vegetatively growing mycelium. A specialized anatomical structure, called a clamp connection, is formed at each hyphal septum. As with the structurally similar hook in the ascomycetes, the clamp connection in the basidiomycetes is required for controlled transfer of nuclei during cell division, to maintain the dikaryotic stage with two genetically different nuclei in each hyphal compartment.[87] A basidiocarp is formed in which club-like structures known as basidia generate haploid basidiospores after karyogamy and meiosis.[88] The most commonly known basidiocarps are mushrooms, but they may also take other forms (see Morphology section).
In fungi formerly classified as Zygomycota, haploid hyphae of two individuals fuse, forming a gametangium, a specialized cell structure that becomes a fertile gamete-producing cell. The gametangium develops into a zygospore, a thick-walled spore formed by the union of gametes. When the zygospore germinates, it undergoes meiosis, generating new haploid hyphae, which may then form asexual sporangiospores. These sporangiospores allow the fungus to rapidly disperse and germinate into new genetically identical haploid fungal mycelia.[89]
Spore dispersal
Both asexual and sexual spores or sporangiospores are often actively dispersed by forcible ejection from their reproductive structures. This ejection ensures exit of the spores from the reproductive structures as well as traveling through the air over long distances.
Specialized mechanical and physiological mechanisms, as well as spore surface structures (such as hydrophobins), enable efficient spore ejection.[90] For example, the structure of the spore-bearing cells in some ascomycete species is such that the buildup of substances affecting cell volume and fluid balance enables the explosive discharge of spores into the air.[91] The forcible discharge of single spores termed ballistospores involves formation of a small drop of water (Buller's drop), which upon contact with the spore leads to its projectile release with an initial acceleration of more than 10,000 g;[92] the net result is that the spore is ejected 0.01–0.02 cm, sufficient distance for it to fall through the gills or pores into the air below.[93] Other fungi, like the puffballs, rely on alternative mechanisms for spore release, such as external mechanical forces. The hydnoid fungi (tooth fungi) produce spores on pendant, tooth-like or spine-like projections.[94] The bird's nest fungi use the force of falling water drops to liberate the spores from cup-shaped fruiting bodies.[95] Another strategy is seen in the stinkhorns, a group of fungi with lively colors and putrid odor that attract insects to disperse their spores.[96]
The most common means of spore dispersal is by wind - species using this form of dispersal often produce dry or hydrophobic spores which do not absorb water and are readily scattered by raindrops, for example. Most of the researched species of fungus are transported by wind.[97][98]
Homothallism
In homothallic sexual reproduction, two haploid nuclei derived from the same individual fuse to form a zygote that can then undergo meiosis. Homothallic fungi include species with an aspergillus-like asexual stage (anamorphs) occurring in numerous different genera,[99] several species of the ascomycete genus Cochliobolus,[100] and the ascomycete Pneumocystis jiroveccii.[101] Heitman[102] reviewed evidence bearing on the evolution of sexual reproduction in the fungi and concluded that the earliest mode of sexual reproduction among eukaryotes was likely homothallism, that is, self-fertile unisexual reproduction.
Other sexual processes
Besides regular sexual reproduction with meiosis, certain fungi, such as those in the genera Penicillium and Aspergillus, may exchange genetic material via parasexual processes, initiated by anastomosis between hyphae and plasmogamy of fungal cells.[103] The frequency and relative importance of parasexual events is unclear and may be lower than other sexual processes. It is known to play a role in intraspecific hybridization[104] and is likely required for hybridization between species, which has been associated with major events in fungal evolution.[105]
Evolution
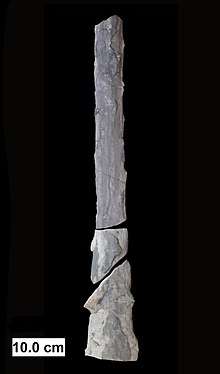
In contrast to plants and animals, the early fossil record of the fungi is meager. Factors that likely contribute to the under-representation of fungal species among fossils include the nature of fungal fruiting bodies, which are soft, fleshy, and easily degradable tissues and the microscopic dimensions of most fungal structures, which therefore are not readily evident. Fungal fossils are difficult to distinguish from those of other microbes, and are most easily identified when they resemble extant fungi.[106] Often recovered from a permineralized plant or animal host, these samples are typically studied by making thin-section preparations that can be examined with light microscopy or transmission electron microscopy.[107] Researchers study compression fossils by dissolving the surrounding matrix with acid and then using light or scanning electron microscopy to examine surface details.[108]
The earliest fossils possessing features typical of fungi date to the Paleoproterozoic era, some 2,400 million years ago (Ma); these multicellular benthic organisms had filamentous structures capable of anastomosis.[109] Other studies (2009) estimate the arrival of fungal organisms at about 760–1060 Ma on the basis of comparisons of the rate of evolution in closely related groups.[110] For much of the Paleozoic Era (542–251 Ma), the fungi appear to have been aquatic and consisted of organisms similar to the extant chytrids in having flagellum-bearing spores.[111] The evolutionary adaptation from an aquatic to a terrestrial lifestyle necessitated a diversification of ecological strategies for obtaining nutrients, including parasitism, saprobism, and the development of mutualistic relationships such as mycorrhiza and lichenization.[112] Recent (2009) studies suggest that the ancestral ecological state of the Ascomycota was saprobism, and that independent lichenization events have occurred multiple times.[113]
In May 2019, scientists reported the discovery of a fossilized fungus, named Ourasphaira giraldae, in the Canadian Arctic, that may have grown on land a billion years ago, well before plants were living on land.[114][115][116] Earlier, it had been presumed that the fungi colonized the land during the Cambrian (542–488.3 Ma), also long before land plants.[117] Fossilized hyphae and spores recovered from the Ordovician of Wisconsin (460 Ma) resemble modern-day Glomerales, and existed at a time when the land flora likely consisted of only non-vascular bryophyte-like plants.[118] Prototaxites, which was probably a fungus or lichen, would have been the tallest organism of the late Silurian and early Devonian. Fungal fossils do not become common and uncontroversial until the early Devonian (416–359.2 Ma), when they occur abundantly in the Rhynie chert, mostly as Zygomycota and Chytridiomycota.[117][119][120] At about this same time, approximately 400 Ma, the Ascomycota and Basidiomycota diverged,[121] and all modern classes of fungi were present by the Late Carboniferous (Pennsylvanian, 318.1–299 Ma).[122]
Lichen-like fossils have been found in the Doushantuo Formation in southern China dating back to 635–551 Ma.[123] Lichens formed a component of the early terrestrial ecosystems, and the estimated age of the oldest terrestrial lichen fossil is 400 Ma;[124] this date corresponds to the age of the oldest known sporocarp fossil, a Paleopyrenomycites species found in the Rhynie Chert.[125] The oldest fossil with microscopic features resembling modern-day basidiomycetes is Palaeoancistrus, found permineralized with a fern from the Pennsylvanian.[126] Rare in the fossil record are the Homobasidiomycetes (a taxon roughly equivalent to the mushroom-producing species of the Agaricomycetes). Two amber-preserved specimens provide evidence that the earliest known mushroom-forming fungi (the extinct species Archaeomarasmius leggetti) appeared during the late Cretaceous, 90 Ma.[127][128]
Some time after the Permian–Triassic extinction event (251.4 Ma), a fungal spike (originally thought to be an extraordinary abundance of fungal spores in sediments) formed, suggesting that fungi were the dominant life form at this time, representing nearly 100% of the available fossil record for this period.[129] However, the relative proportion of fungal spores relative to spores formed by algal species is difficult to assess,[130] the spike did not appear worldwide,[131][132] and in many places it did not fall on the Permian–Triassic boundary.[133]
65 million years ago, immediately after the Cretaceous–Paleogene extinction event that famously killed off most dinosaurs, there is a dramatic increase in evidence of fungi, apparently the death of most plant and animal species leading to a huge fungal bloom like "a massive compost heap".[134]
Taxonomy
Although commonly included in botany curricula and textbooks, fungi are more closely related to animals than to plants and are placed with the animals in the monophyletic group of opisthokonts.[135] Analyses using molecular phylogenetics support a monophyletic origin of fungi.[44] The taxonomy of fungi is in a state of constant flux, especially due to recent research based on DNA comparisons. These current phylogenetic analyses often overturn classifications based on older and sometimes less discriminative methods based on morphological features and biological species concepts obtained from experimental matings.[136]
There is no unique generally accepted system at the higher taxonomic levels and there are frequent name changes at every level, from species upwards. Efforts among researchers are now underway to establish and encourage usage of a unified and more consistent nomenclature.[44][137] Fungal species can also have multiple scientific names depending on their life cycle and mode (sexual or asexual) of reproduction. Web sites such as Index Fungorum and ITIS list current names of fungal species (with cross-references to older synonyms).
The 2007 classification of Kingdom Fungi is the result of a large-scale collaborative research effort involving dozens of mycologists and other scientists working on fungal taxonomy.[44] It recognizes seven phyla, two of which—the Ascomycota and the Basidiomycota—are contained within a branch representing subkingdom Dikarya, the most species rich and familiar group, including all the mushrooms, most food-spoilage molds, most plant pathogenic fungi, and the beer, wine, and bread yeasts. The accompanying cladogram depicts the major fungal taxa and their relationship to opisthokont and unikont organisms, based on the work of Philippe Silar,[138] "The Mycota: A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research"[139] and Tedersoo et al. 2018.[140] The lengths of the branches are not proportional to evolutionary distances.
| Zoosporia |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Basidiomycota |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ascomycota |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Taxonomic groups
.png)
The major phyla (sometimes called divisions) of fungi have been classified mainly on the basis of characteristics of their sexual reproductive structures. Currently, seven phyla are proposed: Microsporidia, Chytridiomycota, Blastocladiomycota, Neocallimastigomycota, Glomeromycota, Ascomycota, and Basidiomycota.[44]
Phylogenetic analysis has demonstrated that the Microsporidia, unicellular parasites of animals and protists, are fairly recent and highly derived endobiotic fungi (living within the tissue of another species).[111][141] One 2006 study concludes that the Microsporidia are a sister group to the true fungi; that is, they are each other's closest evolutionary relative.[142] Hibbett and colleagues suggest that this analysis does not clash with their classification of the Fungi, and although the Microsporidia are elevated to phylum status, it is acknowledged that further analysis is required to clarify evolutionary relationships within this group.[44]
The Chytridiomycota are commonly known as chytrids. These fungi are distributed worldwide. Chytrids and their close relatives Neocallimastigomycota and Blastocladiomycota (below) are the only fungi with active motility, producing zoospores that are capable of active movement through aqueous phases with a single flagellum, leading early taxonomists to classify them as protists. Molecular phylogenies, inferred from rRNA sequences in ribosomes, suggest that the Chytrids are a basal group divergent from the other fungal phyla, consisting of four major clades with suggestive evidence for paraphyly or possibly polyphyly.[143]
The Blastocladiomycota were previously considered a taxonomic clade within the Chytridiomycota. Recent molecular data and ultrastructural characteristics, however, place the Blastocladiomycota as a sister clade to the Zygomycota, Glomeromycota, and Dikarya (Ascomycota and Basidiomycota). The blastocladiomycetes are saprotrophs, feeding on decomposing organic matter, and they are parasites of all eukaryotic groups. Unlike their close relatives, the chytrids, most of which exhibit zygotic meiosis, the blastocladiomycetes undergo sporic meiosis.[111]
The Neocallimastigomycota were earlier placed in the phylum Chytridomycota. Members of this small phylum are anaerobic organisms, living in the digestive system of larger herbivorous mammals and in other terrestrial and aquatic environments enriched in cellulose (e.g., domestic waste landfill sites).[144] They lack mitochondria but contain hydrogenosomes of mitochondrial origin. As in the related chrytrids, neocallimastigomycetes form zoospores that are posteriorly uniflagellate or polyflagellate.[44]

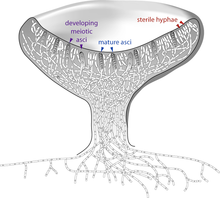
Members of the Glomeromycota form arbuscular mycorrhizae, a form of mutualist symbiosis wherein fungal hyphae invade plant root cells and both species benefit from the resulting increased supply of nutrients. All known Glomeromycota species reproduce asexually.[81] The symbiotic association between the Glomeromycota and plants is ancient, with evidence dating to 400 million years ago.[145] Formerly part of the Zygomycota (commonly known as 'sugar' and 'pin' molds), the Glomeromycota were elevated to phylum status in 2001 and now replace the older phylum Zygomycota.[146] Fungi that were placed in the Zygomycota are now being reassigned to the Glomeromycota, or the subphyla incertae sedis Mucoromycotina, Kickxellomycotina, the Zoopagomycotina and the Entomophthoromycotina.[44] Some well-known examples of fungi formerly in the Zygomycota include black bread mold (Rhizopus stolonifer), and Pilobolus species, capable of ejecting spores several meters through the air.[147] Medically relevant genera include Mucor, Rhizomucor, and Rhizopus.
The Ascomycota, commonly known as sac fungi or ascomycetes, constitute the largest taxonomic group within the Eumycota.[43] These fungi form meiotic spores called ascospores, which are enclosed in a special sac-like structure called an ascus. This phylum includes morels, a few mushrooms and truffles, unicellular yeasts (e.g., of the genera Saccharomyces, Kluyveromyces, Pichia, and Candida), and many filamentous fungi living as saprotrophs, parasites, and mutualistic symbionts (e.g. lichens). Prominent and important genera of filamentous ascomycetes include Aspergillus, Penicillium, Fusarium, and Claviceps. Many ascomycete species have only been observed undergoing asexual reproduction (called anamorphic species), but analysis of molecular data has often been able to identify their closest teleomorphs in the Ascomycota.[148] Because the products of meiosis are retained within the sac-like ascus, ascomycetes have been used for elucidating principles of genetics and heredity (e.g., Neurospora crassa).[149]
Members of the Basidiomycota, commonly known as the club fungi or basidiomycetes, produce meiospores called basidiospores on club-like stalks called basidia. Most common mushrooms belong to this group, as well as rust and smut fungi, which are major pathogens of grains. Other important basidiomycetes include the maize pathogen Ustilago maydis,[150] human commensal species of the genus Malassezia,[151] and the opportunistic human pathogen, Cryptococcus neoformans.[152]
Fungus-like organisms
Because of similarities in morphology and lifestyle, the slime molds (mycetozoans, plasmodiophorids, acrasids, Fonticula and labyrinthulids, now in Amoebozoa, Rhizaria, Excavata, Opisthokonta and Stramenopiles, respectively), water molds (oomycetes) and hyphochytrids (both Stramenopiles) were formerly classified in the kingdom Fungi, in groups like Mastigomycotina, Gymnomycota and Phycomycetes. The slime molds were studied also as protozoans, leading to an ambiregnal, duplicated taxonomy.
Unlike true fungi, the cell walls of oomycetes contain cellulose and lack chitin. Hyphochytrids have both chitin and cellulose. Slime molds lack a cell wall during the assimilative phase (except labyrinthulids, which have a wall of scales), and ingest nutrients by ingestion (phagocytosis, except labyrinthulids) rather than absorption (osmotrophy, as fungi, labyrinthulids, oomycetes and hyphochytrids). Neither water molds nor slime molds are closely related to the true fungi, and, therefore, taxonomists no longer group them in the kingdom Fungi. Nonetheless, studies of the oomycetes and myxomycetes are still often included in mycology textbooks and primary research literature.[153]
The Eccrinales and Amoebidiales are opisthokont protists, previously thought to be zygomycete fungi. Other groups now in Opisthokonta (e.g., Corallochytrium, Ichthyosporea) were also at given time classified as fungi. The genus Blastocystis, now in Stramenopiles, was originally classified as a yeast. Ellobiopsis, now in Alveolata, was considered a chytrid. The bacteria were also included in fungi in some classifications, as the group Schizomycetes.
The Rozellida clade, including the "ex-chytrid" Rozella, is a genetically disparate group known mostly from environmental DNA sequences that is a sister group to fungi. Members of the group that have been isolated lack the chitinous cell wall that is characteristic of fungi.
The nucleariids may be the next sister group to the eumycete clade, and as such could be included in an expanded fungal kingdom.[135] Many Actinomycetales (Actinobacteria), a group with many filamentous bacteria, were also long believed to be fungi.[154][155]
Ecology

Although often inconspicuous, fungi occur in every environment on Earth and play very important roles in most ecosystems. Along with bacteria, fungi are the major decomposers in most terrestrial (and some aquatic) ecosystems, and therefore play a critical role in biogeochemical cycles[156] and in many food webs. As decomposers, they play an essential role in nutrient cycling, especially as saprotrophs and symbionts, degrading organic matter to inorganic molecules, which can then re-enter anabolic metabolic pathways in plants or other organisms.[157][158]
Symbiosis
Many fungi have important symbiotic relationships with organisms from most if not all Kingdoms.[159][160][161] These interactions can be mutualistic or antagonistic in nature, or in the case of commensal fungi are of no apparent benefit or detriment to the host.[162][163][164]
With plants
Mycorrhizal symbiosis between plants and fungi is one of the most well-known plant–fungus associations and is of significant importance for plant growth and persistence in many ecosystems; over 90% of all plant species engage in mycorrhizal relationships with fungi and are dependent upon this relationship for survival.[165]
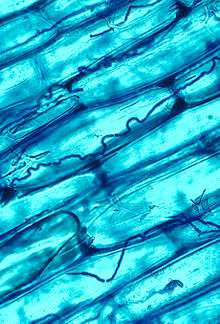
The mycorrhizal symbiosis is ancient, dating to at least 400 million years ago.[145] It often increases the plant's uptake of inorganic compounds, such as nitrate and phosphate from soils having low concentrations of these key plant nutrients.[157][166] The fungal partners may also mediate plant-to-plant transfer of carbohydrates and other nutrients.[167] Such mycorrhizal communities are called "common mycorrhizal networks".[168][169] A special case of mycorrhiza is myco-heterotrophy, whereby the plant parasitizes the fungus, obtaining all of its nutrients from its fungal symbiont.[170] Some fungal species inhabit the tissues inside roots, stems, and leaves, in which case they are called endophytes.[171] Similar to mycorrhiza, endophytic colonization by fungi may benefit both symbionts; for example, endophytes of grasses impart to their host increased resistance to herbivores and other environmental stresses and receive food and shelter from the plant in return.[172]
With algae and cyanobacteria

Lichens are a symbiotic relationship between fungi and photosynthetic algae or cyanobacteria. The photosynthetic partner in the relationship is referred to in lichen terminology as a "photobiont". The fungal part of the relationship is composed mostly of various species of ascomycetes and a few basidiomycetes.[173] Lichens occur in every ecosystem on all continents, play a key role in soil formation and the initiation of biological succession,[174] and are prominent in some extreme environments, including polar, alpine, and semiarid desert regions.[175] They are able to grow on inhospitable surfaces, including bare soil, rocks, tree bark, wood, shells, barnacles and leaves.[176] As in mycorrhizas, the photobiont provides sugars and other carbohydrates via photosynthesis to the fungus, while the fungus provides minerals and water to the photobiont. The functions of both symbiotic organisms are so closely intertwined that they function almost as a single organism; in most cases the resulting organism differs greatly from the individual components. Lichenization is a common mode of nutrition for fungi; around 20% of fungi—between 17,500 and 20,000 described species—are lichenized.[177] Characteristics common to most lichens include obtaining organic carbon by photosynthesis, slow growth, small size, long life, long-lasting (seasonal) vegetative reproductive structures, mineral nutrition obtained largely from airborne sources, and greater tolerance of desiccation than most other photosynthetic organisms in the same habitat.[178]
With insects
Many insects also engage in mutualistic relationships with fungi. Several groups of ants cultivate fungi in the order Agaricales as their primary food source, while ambrosia beetles cultivate various species of fungi in the bark of trees that they infest.[179] Likewise, females of several wood wasp species (genus Sirex) inject their eggs together with spores of the wood-rotting fungus Amylostereum areolatum into the sapwood of pine trees; the growth of the fungus provides ideal nutritional conditions for the development of the wasp larvae.[180] At least one species of stingless bee has a relationship with a fungus in the genus Monascus, where the larvae consume and depend on fungus transferred from old to new nests.[181] Termites on the African savannah are also known to cultivate fungi,[159] and yeasts of the genera Candida and Lachancea inhabit the gut of a wide range of insects, including neuropterans, beetles, and cockroaches; it is not known whether these fungi benefit their hosts.[182]Fungi ingrowing dead wood are essential for xylophagous insects (e.g. woodboring beetles).[183][184][185] They deliver nutrients needed by xylophages to nutritionally scarce dead wood.[186][184][185] Thanks to this nutritional enrichment the larvae of woodboring insect is able to grow and develop to adulthood.[183] The larvae of many families of fungicolous flies, particularly those within the superfamily Sciaroidea such as the Mycetophilidae and some Keroplatidae feed on fungal fruiting bodies and sterile mycorrhizae.[187]
As pathogens and parasites

Many fungi are parasites on plants, animals (including humans), and other fungi. Serious pathogens of many cultivated plants causing extensive damage and losses to agriculture and forestry include the rice blast fungus Magnaporthe oryzae,[188] tree pathogens such as Ophiostoma ulmi and Ophiostoma novo-ulmi causing Dutch elm disease[189] and Cryphonectria parasitica responsible for chestnut blight,[190] and plant pathogens in the genera Fusarium, Ustilago, Alternaria, and Cochliobolus.[163] Some carnivorous fungi, like Paecilomyces lilacinus, are predators of nematodes, which they capture using an array of specialized structures such as constricting rings or adhesive nets.[191] Many fungi that are plant pathogens, such as Magnaporthe oryzae, can switch from being biotrophic (parasitic on living plants) to being necrotrophic (feeding on the dead tissues of plants they have killed).[192] This same principle is applied to fungi-feeding parasites, including Asterotremella albida, which feeds on the fruit bodies of other fungi both while they are living and after they are dead.[193]
Some fungi can cause serious diseases in humans, several of which may be fatal if untreated. These include aspergillosis, candidiasis, coccidioidomycosis, cryptococcosis, histoplasmosis, mycetomas, and paracoccidioidomycosis. Furthermore, persons with immuno-deficiencies are particularly susceptible to disease by genera such as Aspergillus, Candida, Cryptoccocus,[164][194][195] Histoplasma,[196] and Pneumocystis.[197] Other fungi can attack eyes, nails, hair, and especially skin, the so-called dermatophytic and keratinophilic fungi, and cause local infections such as ringworm and athlete's foot.[198] Fungal spores are also a cause of allergies, and fungi from different taxonomic groups can evoke allergic reactions.[199]
As targets of mycoparasites
The organisms which parasitize fungi are known as mycoparasitic organisms. Certain species of the genus Pythium, which are oomycetes, have potential as biocontrol agents against certain fungi.[200] Fungi can also act as mycoparasites or antagonists of other fungi, such as Hypomyces chrysospermus, which grows on bolete mushrooms. Fungi can also become the target of infection by mycoviruses.[201][202]
Mycotoxins
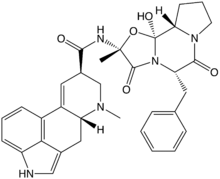
Many fungi produce biologically active compounds, several of which are toxic to animals or plants and are therefore called mycotoxins. Of particular relevance to humans are mycotoxins produced by molds causing food spoilage, and poisonous mushrooms (see above). Particularly infamous are the lethal amatoxins in some Amanita mushrooms, and ergot alkaloids, which have a long history of causing serious epidemics of ergotism (St Anthony's Fire) in people consuming rye or related cereals contaminated with sclerotia of the ergot fungus, Claviceps purpurea.[203] Other notable mycotoxins include the aflatoxins, which are insidious liver toxins and highly carcinogenic metabolites produced by certain Aspergillus species often growing in or on grains and nuts consumed by humans, ochratoxins, patulin, and trichothecenes (e.g., T-2 mycotoxin) and fumonisins, which have significant impact on human food supplies or animal livestock.[204]
Mycotoxins are secondary metabolites (or natural products), and research has established the existence of biochemical pathways solely for the purpose of producing mycotoxins and other natural products in fungi.[33] Mycotoxins may provide fitness benefits in terms of physiological adaptation, competition with other microbes and fungi, and protection from consumption (fungivory).[205][206] Many fungal secondary metabolites (or derivatives) are used medically, as described under Human Use below.
Pathogenic mechanisms
Ustilago maydis is a pathogenic plant fungus that causes smut disease in maize and teosinte. Plants have evolved efficient defense systems against pathogenic microbes such as U. maydis. A rapid defense reaction after pathogen attack is the oxidative burst where the plant produces reactive oxygen species at the site of the attempted invasion. U. maydis can respond to the oxidative burst with an oxidative stress response, regulated by the gene YAP1. The response protects U. maydis from the host defense, and is necessary for the pathogen's virulence.[207] Furthermore, U. maydis has a well-established recombinational DNA repair system which acts during mitosis and meiosis.[208] The system may assist the pathogen in surviving DNA damage arising from the host plant's oxidative defensive response to infection.[209]
Cryptococcus neoformans is an encapsulated yeast that can live in both plants and animals. C. neoformans usually infects the lungs, where it is phagocytosed by alveolar macrophages.[210] Some C. neoformans can survive inside macrophages, which appears to be the basis for latency, disseminated disease, and resistance to antifungal agents. One mechanism by which C. neoformans survives the hostile macrophage environment is by up-regulating the expression of genes involved in the oxidative stress response.[210] Another mechanism involves meiosis. The majority of C. neoformans are mating "type a". Filaments of mating "type a" ordinarily have haploid nuclei, but they can become diploid (perhaps by endoduplication or by stimulated nuclear fusion) to form blastospores. The diploid nuclei of blastospores can undergo meiosis, including recombination, to form haploid basidiospores that can be dispersed.[211] This process is referred to as monokaryotic fruiting. This process requires a gene called DMC1, which is a conserved homologue of genes recA in bacteria and RAD51 in eukaryotes, that mediates homologous chromosome pairing during meiosis and repair of DNA double-strand breaks. Thus, C. neoformans can undergo a meiosis, monokaryotic fruiting, that promotes recombinational repair in the oxidative, DNA damaging environment of the host macrophage, and the repair capability may contribute to its virulence.[209][211]
Human use
The human use of fungi for food preparation or preservation and other purposes is extensive and has a long history. Mushroom farming and mushroom gathering are large industries in many countries. The study of the historical uses and sociological impact of fungi is known as ethnomycology. Because of the capacity of this group to produce an enormous range of natural products with antimicrobial or other biological activities, many species have long been used or are being developed for industrial production of antibiotics, vitamins, and anti-cancer and cholesterol-lowering drugs. More recently, methods have been developed for genetic engineering of fungi,[212] enabling metabolic engineering of fungal species. For example, genetic modification of yeast species[213]—which are easy to grow at fast rates in large fermentation vessels—has opened up ways of pharmaceutical production that are potentially more efficient than production by the original source organisms.[214]
Therapeutic uses
Modern chemotherapeutics
Many species produce metabolites that are major sources of pharmacologically active drugs. Particularly important are the antibiotics, including the penicillins, a structurally related group of β-lactam antibiotics that are synthesized from small peptides. Although naturally occurring penicillins such as penicillin G (produced by Penicillium chrysogenum) have a relatively narrow spectrum of biological activity, a wide range of other penicillins can be produced by chemical modification of the natural penicillins. Modern penicillins are semisynthetic compounds, obtained initially from fermentation cultures, but then structurally altered for specific desirable properties.[215] Other antibiotics produced by fungi include: ciclosporin, commonly used as an immunosuppressant during transplant surgery; and fusidic acid, used to help control infection from methicillin-resistant Staphylococcus aureus bacteria.[216] Widespread use of antibiotics for the treatment of bacterial diseases, such as tuberculosis, syphilis, leprosy, and others began in the early 20th century and continues to date. In nature, antibiotics of fungal or bacterial origin appear to play a dual role: at high concentrations they act as chemical defense against competition with other microorganisms in species-rich environments, such as the rhizosphere, and at low concentrations as quorum-sensing molecules for intra- or interspecies signaling.[217] Other drugs produced by fungi include griseofulvin isolated from Penicillium griseofulvum, used to treat fungal infections,[218] and statins (HMG-CoA reductase inhibitors), used to inhibit cholesterol synthesis. Examples of statins found in fungi include mevastatin from Penicillium citrinum and lovastatin from Aspergillus terreus and the oyster mushroom.[219] Fungi produce compounds that inhibit viruses[220][221] and cancer cells.[222][223] Specific metabolites, such as polysaccharide-K, ergotamine, and β-lactam antibiotics, are routinely used in clinical medicine. The shiitake mushroom is a source of lentinan, a clinical drug approved for use in cancer treatments in several countries, including Japan.[224][225] In Europe and Japan, polysaccharide-K (brand name Krestin), a chemical derived from Trametes versicolor, is an approved adjuvant for cancer therapy.[226]
Traditional and folk medicine


Certain mushrooms enjoy usage as therapeutics in folk medicines, such as Traditional Chinese medicine. Notable medicinal mushrooms with a well-documented history of use include Agaricus subrufescens,[222][227] Ganoderma lucidum,[228] Psilocybe and Ophiocordyceps sinensis.[229]
Cultured foods
Baker's yeast or Saccharomyces cerevisiae, a unicellular fungus, is used to make bread and other wheat-based products, such as pizza dough and dumplings.[230] Yeast species of the genus Saccharomyces are also used to produce alcoholic beverages through fermentation.[231] Shoyu koji mold (Aspergillus oryzae) is an essential ingredient in brewing Shoyu (soy sauce) and sake, and the preparation of miso,[232] while Rhizopus species are used for making tempeh.[233] Several of these fungi are domesticated species that were bred or selected according to their capacity to ferment food without producing harmful mycotoxins (see below), which are produced by very closely related Aspergilli.[234] Quorn, a meat substitute, is made from Fusarium venenatum.[235]
In food

Edible mushrooms include commercially raised and wild-harvested fungi. Agaricus bisporus, sold as button mushrooms when small or Portobello mushrooms when larger, is the most widely cultivated species in the West, used in salads, soups, and many other dishes. Many Asian fungi are commercially grown and have increased in popularity in the West. They are often available fresh in grocery stores and markets, including straw mushrooms (Volvariella volvacea), oyster mushrooms (Pleurotus ostreatus), shiitakes (Lentinula edodes), and enokitake (Flammulina spp.).[236]

Many other mushroom species are harvested from the wild for personal consumption or commercial sale. Milk mushrooms, morels, chanterelles, truffles, black trumpets, and porcini mushrooms (Boletus edulis) (also known as king boletes) demand a high price on the market. They are often used in gourmet dishes.[237]
Certain types of cheeses require inoculation of milk curds with fungal species that impart a unique flavor and texture to the cheese. Examples include the blue color in cheeses such as Stilton or Roquefort, which are made by inoculation with Penicillium roqueforti.[238] Molds used in cheese production are non-toxic and are thus safe for human consumption; however, mycotoxins (e.g., aflatoxins, roquefortine C, patulin, or others) may accumulate because of growth of other fungi during cheese ripening or storage.[239]
Poisonous fungi
Many mushroom species are poisonous to humans and cause a range of reactions including slight digestive problems, allergic reactions, hallucinations, severe organ failure, and death. Genera with mushrooms containing deadly toxins include Conocybe, Galerina, Lepiota, and, the most infamous, Amanita.[240] The latter genus includes the destroying angel (A. virosa) and the death cap (A. phalloides), the most common cause of deadly mushroom poisoning.[241] The false morel (Gyromitra esculenta) is occasionally considered a delicacy when cooked, yet can be highly toxic when eaten raw.[242] Tricholoma equestre was considered edible until it was implicated in serious poisonings causing rhabdomyolysis.[243] Fly agaric mushrooms (Amanita muscaria) also cause occasional non-fatal poisonings, mostly as a result of ingestion for its hallucinogenic properties. Historically, fly agaric was used by different peoples in Europe and Asia and its present usage for religious or shamanic purposes is reported from some ethnic groups such as the Koryak people of north-eastern Siberia.[244]
As it is difficult to accurately identify a safe mushroom without proper training and knowledge, it is often advised to assume that a wild mushroom is poisonous and not to consume it.[245][246]
Pest control

In agriculture, fungi may be useful if they actively compete for nutrients and space with pathogenic microorganisms such as bacteria or other fungi via the competitive exclusion principle,[247] or if they are parasites of these pathogens. For example, certain species may be used to eliminate or suppress the growth of harmful plant pathogens, such as insects, mites, weeds, nematodes, and other fungi that cause diseases of important crop plants.[248] This has generated strong interest in practical applications that use these fungi in the biological control of these agricultural pests. Entomopathogenic fungi can be used as biopesticides, as they actively kill insects.[249] Examples that have been used as biological insecticides are Beauveria bassiana, Metarhizium spp, Hirsutella spp, Paecilomyces (Isaria) spp, and Lecanicillium lecanii.[250][251] Endophytic fungi of grasses of the genus Neotyphodium, such as N. coenophialum, produce alkaloids that are toxic to a range of invertebrate and vertebrate herbivores. These alkaloids protect grass plants from herbivory, but several endophyte alkaloids can poison grazing animals, such as cattle and sheep.[252] Infecting cultivars of pasture or forage grasses with Neotyphodium endophytes is one approach being used in grass breeding programs; the fungal strains are selected for producing only alkaloids that increase resistance to herbivores such as insects, while being non-toxic to livestock.[253][254]
Bioremediation
Certain fungi, in particular white-rot fungi, can degrade insecticides, herbicides, pentachlorophenol, creosote, coal tars, and heavy fuels and turn them into carbon dioxide, water, and basic elements.[255] Fungi have been shown to biomineralize uranium oxides, suggesting they may have application in the bioremediation of radioactively polluted sites.[256][257][258]
Model organisms
Several pivotal discoveries in biology were made by researchers using fungi as model organisms, that is, fungi that grow and sexually reproduce rapidly in the laboratory. For example, the one gene-one enzyme hypothesis was formulated by scientists using the bread mold Neurospora crassa to test their biochemical theories.[259] Other important model fungi are Aspergillus nidulans and the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, each of which with a long history of use to investigate issues in eukaryotic cell biology and genetics, such as cell cycle regulation, chromatin structure, and gene regulation. Other fungal models have more recently emerged that address specific biological questions relevant to medicine, plant pathology, and industrial uses; examples include Candida albicans, a dimorphic, opportunistic human pathogen,[260] Magnaporthe grisea, a plant pathogen,[261] and Pichia pastoris, a yeast widely used for eukaryotic protein production.[262]
Others
Fungi are used extensively to produce industrial chemicals like citric, gluconic, lactic, and malic acids,[263] and industrial enzymes, such as lipases used in biological detergents,[264] cellulases used in making cellulosic ethanol[265] and stonewashed jeans,[266] and amylases,[267] invertases, proteases and xylanases.[268]
See also
- Conservation of fungi
- DPVweb
- Marine fungi
- MycoBank
- Mycosis
- Outline of fungi
- Plant pathology
References
- Moore RT (1980). "Taxonomic proposals for the classification of marine yeasts and other yeast-like fungi including the smuts". Botanica Marina. 23: 361–373.
- The classification system presented here is based on the 2007 phylogenetic study by Hibbett et al.
- /ˈfʌndʒaɪ/ (




- "Fungus". Oxford Dictionaries. Retrieved 26 February 2011.
- Hawksworth DL, Lücking R (July 2017). Fungal Diversity Revisited: 2.2 to 3.8 Million Species. Microbiology Spectrum. 5. pp. 79–95. doi:10.1128/microbiolspec.FUNK-0052-2016. ISBN 978-1-55581-957-6. PMID 28752818.
- "Stop neglecting fungi". Nature Microbiology. 2 (8): 17120. 25 July 2017. doi:10.1038/nmicrobiol.2017.120. PMID 28741610.
- Simpson DP (1979). Cassell's Latin Dictionary (5 ed.). London, UK: Cassell Ltd. p. 883. ISBN 978-0-304-52257-6.
- Ainsworth, p. 2.
- Mitzka W, ed. (1960). Etymologisches Wörterbuch der deutschen Sprache. Berlin: Walter de Gruyter.
- Alexopoulos et al., p. 1.
- Persoon, Christiaan Hendrik (1796). Observationes Mycologicae (in Latin). Part 1. Leipzig, (Germany): Peter Philipp Wolf.
- Greville, Robert Kaye (1824). Scottish Cryptogamie Flora: Or Coloured Figures and Descriptions of Cryptogamic Plants, Belonging Chiefly to the Order Fungi. vol. 2. Edinburgh, Scotland: Maclachland and Stewart. p. 65. From p. 65: "This little plant will probably not prove rare in Great Britain, when mycology shall be more studied."
- Smith, James Edward; Hooker, William Jackson, ed. (1836). Berkeley, Miles Joseph (ed.). The English Flora of Sir James Edward Smith. vol. 5, part II: "Class XXIV. Cryptogamia". London, England: Longman, Rees, Orme, Brown, Green & Longman. p. 7.CS1 maint: extra text: authors list (link) From p. 7: "This has arisen, I conceive, partly from the practical difficulty of preserving specimens for the herbarium, partly from the absence of any general work, adapted to the immense advances which have of late years been made in the study of Mycology."
- "LIAS Glossary". Retrieved 14 August 2013.
- Bruns T (October 2006). "Evolutionary biology: a kingdom revised". Nature. 443 (7113): 758–61. Bibcode:2006Natur.443..758B. doi:10.1038/443758a. PMID 17051197.
- Baldauf SL, Palmer JD (December 1993). "Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins". Proceedings of the National Academy of Sciences of the United States of America. 90 (24): 11558–62. Bibcode:1993PNAS...9011558B. doi:10.1073/pnas.90.24.11558. PMC 48023. PMID 8265589.
- Deacon, p. 4.
- Deacon, pp. 128–129.
- Alexopoulos et al., pp. 28–33.
- Alexopoulos et al., pp. 31–32.
- Shoji JY, Arioka M, Kitamoto K (2006). "Possible involvement of pleiomorphic vacuolar networks in nutrient recycling in filamentous fungi". Autophagy. 2 (3): 226–7. doi:10.4161/auto.2695. PMID 16874107.
- Deacon, p. 58.
- Zabriskie TM, Jackson MD (February 2000). "Lysine biosynthesis and metabolism in fungi". Natural Product Reports. 17 (1): 85–97. doi:10.1039/a801345d. PMID 10714900.
- Xu H, Andi B, Qian J, West AH, Cook PF (2006). "The alpha-aminoadipate pathway for lysine biosynthesis in fungi". Cell Biochemistry and Biophysics. 46 (1): 43–64. doi:10.1385/CBB:46:1:43. PMID 16943623.
- Alexopoulos et al., pp. 27–28.
- Alexopoulos et al., p. 685.
- Alexopoulos et al., p. 30.
- Desjardin DE, Perry BA, Lodge DJ, Stevani CV, Nagasawa E (2010). "Luminescent Mycena: new and noteworthy species". Mycologia. 102 (2): 459–77. doi:10.3852/09-197. PMID 20361513.
- Alexopoulos et al., pp. 32–33.
- Bowman SM, Free SJ (August 2006). "The structure and synthesis of the fungal cell wall". BioEssays. 28 (8): 799–808. doi:10.1002/bies.20441. PMID 16927300.
- Alexopoulos et al., p. 33.
- Mihail JD, Bruhn JN (November 2005). "Foraging behaviour of Armillaria rhizomorph systems". Mycological Research. 109 (Pt 11): 1195–207. doi:10.1017/S0953756205003606. PMID 16279413.
- Keller NP, Turner G, Bennett JW (December 2005). "Fungal secondary metabolism - from biochemistry to genomics". Nature Reviews. Microbiology. 3 (12): 937–47. doi:10.1038/nrmicro1286. PMID 16322742.
- Wu S, Schalk M, Clark A, Miles RB, Coates R, Chappell J (November 2006). "Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants". Nature Biotechnology. 24 (11): 1441–7. doi:10.1038/nbt1251. PMID 17057703.
- Tudzynski B (March 2005). "Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology". Applied Microbiology and Biotechnology. 66 (6): 597–611. doi:10.1007/s00253-004-1805-1. PMID 15578178.
- Vaupotic T, Veranic P, Jenoe P, Plemenitas A (June 2008). "Mitochondrial mediation of environmental osmolytes discrimination during osmoadaptation in the extremely halotolerant black yeast Hortaea werneckii". Fungal Genetics and Biology. 45 (6): 994–1007. doi:10.1016/j.fgb.2008.01.006. PMID 18343697.
- Dadachova E, Bryan RA, Huang X, Moadel T, Schweitzer AD, Aisen P, Nosanchuk JD, Casadevall A (2007). "Ionizing radiation changes the electronic properties of melanin and enhances the growth of melanized fungi". PLOS ONE. 2 (5): e457. Bibcode:2007PLoSO...2..457D. doi:10.1371/journal.pone.0000457. PMC 1866175. PMID 17520016.

- Raghukumar C, Raghukumar S (1998). "Barotolerance of fungi isolated from deep-sea sediments of the Indian Ocean". Aquatic Microbial Ecology. 15 (2): 153–163. doi:10.3354/ame015153.
- Sancho LG, de la Torre R, Horneck G, Ascaso C, de Los Rios A, Pintado A, Wierzchos J, Schuster M (June 2007). "Lichens survive in space: results from the 2005 LICHENS experiment". Astrobiology. 7 (3): 443–54. Bibcode:2007AsBio...7..443S. doi:10.1089/ast.2006.0046. PMID 17630840.
- Brem FM, Lips KR (September 2008). "Batrachochytrium dendrobatidis infection patterns among Panamanian amphibian species, habitats and elevations during epizootic and enzootic stages". Diseases of Aquatic Organisms. 81 (3): 189–202. doi:10.3354/dao01960. PMID 18998584.
- Le Calvez T, Burgaud G, Mahé S, Barbier G, Vandenkoornhuyse P (October 2009). "Fungal diversity in deep-sea hydrothermal ecosystems". Applied and Environmental Microbiology. 75 (20): 6415–21. doi:10.1128/AEM.00653-09. PMC 2765129. PMID 19633124.
- Mueller GM, Schmit JP (2006). "Fungal biodiversity: what do we know? What can we predict?". Biodiversity and Conservation. 16: 1–5. doi:10.1007/s10531-006-9117-7.
- Kirk et al., p. 489.
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, et al. (May 2007). "A higher-level phylogenetic classification of the Fungi" (PDF). Mycological Research. 111 (Pt 5): 509–47. CiteSeerX 10.1.1.626.9582. doi:10.1016/j.mycres.2007.03.004. PMID 17572334. Archived from the original (PDF) on 26 March 2009.
- According to one 2001 estimate, some 10,000 fungal diseases are known. Struck C (2006). "Infection strategies of plant parasitic fungi". In Cooke BM, Jones DG, Kaye B (eds.). The Epidemiology of Plant Diseases. Berlin, Germany: Springer. p. 117. ISBN 978-1-4020-4580-6.
- Peintner U, Pöder R, Pümpel T (1998). "The Iceman's fungi". Mycological Research. 102 (10): 1153–1162. doi:10.1017/S0953756298006546.
- Ainsworth, p. 1.
- Alexopoulos et al., pp. 1–2.
- Ainsworth, p. 18.
- Hawksworth DL (September 2006). "Pandora's mycological box: molecular sequences vs. morphology in understanding fungal relationships and biodiversity". Revista Iberoamericana de MicologíA. 23 (3): 127–33. doi:10.1016/S1130-1406(06)70031-6. PMID 17196017.
- Harris SD (2008). "Branching of fungal hyphae: regulation, mechanisms and comparison with other branching systems". Mycologia. 100 (6): 823–32. doi:10.3852/08-177. PMID 19202837.
- Deacon, p. 51.
- Deacon, p. 57.
- Chang S-T, Miles PG (2004). Mushrooms: Cultivation, Nutritional Value, Medicinal Effect and Environmental Impact. Boca Raton, Florida: CRC Press. ISBN 978-0-8493-1043-0.
- Parniske M (October 2008). "Arbuscular mycorrhiza: the mother of plant root endosymbioses". Nature Reviews. Microbiology. 6 (10): 763–75. doi:10.1038/nrmicro1987. PMID 18794914.
- Steenkamp ET, Wright J, Baldauf SL (January 2006). "The protistan origins of animals and fungi". Molecular Biology and Evolution. 23 (1): 93–106. doi:10.1093/molbev/msj011. PMID 16151185.
- Stevens DA, Ichinomiya M, Koshi Y, Horiuchi H (September 2006). "Escape of Candida from caspofungin inhibition at concentrations above the MIC (paradoxical effect) accomplished by increased cell wall chitin; evidence for beta-1,6-glucan synthesis inhibition by caspofungin". Antimicrobial Agents and Chemotherapy. 50 (9): 3160–1. doi:10.1128/AAC.00563-06. PMC 1563524. PMID 16940118.
- Hanson, pp. 127–141.
- Ferguson BA, Dreisbach TA, Parks CG, Filip GM, Schmitt CL (2003). "Coarse-scale population structure of pathogenic Armillaria species in a mixed-conifer forest in the Blue Mountains of northeast Oregon". Canadian Journal of Forest Research. 33 (4): 612–623. doi:10.1139/x03-065.
- Alexopoulos et al., pp. 204–205.
- Moss ST (1986). The Biology of Marine Fungi. Cambridge, UK: Cambridge University Press. p. 76. ISBN 978-0-521-30899-1.
- Peñalva MA, Arst HN (September 2002). "Regulation of gene expression by ambient pH in filamentous fungi and yeasts". Microbiology and Molecular Biology Reviews. 66 (3): 426–46, table of contents. doi:10.1128/MMBR.66.3.426-446.2002. PMC 120796. PMID 12208998.
- Howard RJ, Ferrari MA, Roach DH, Money NP (December 1991). "Penetration of hard substrates by a fungus employing enormous turgor pressures". Proceedings of the National Academy of Sciences of the United States of America. 88 (24): 11281–4. Bibcode:1991PNAS...8811281H. doi:10.1073/pnas.88.24.11281. PMC 53118. PMID 1837147.
- Money NP (1998). "Mechanics of invasive fungal growth and the significance of turgor in plant infection". Molecular Genetics of Host-Specific Toxins in Plant Disease: Proceedings of the 3rd Tottori International Symposium on Host-Specific Toxins, Daisen, Tottori, Japan, August 24–29, 1997. Netherlands: Kluwer Academic Publishers. pp. 261–271. ISBN 978-0-7923-4981-5.
- Wang ZY, Jenkinson JM, Holcombe LJ, Soanes DM, Veneault-Fourrey C, Bhambra GK, Talbot NJ (April 2005). "The molecular biology of appressorium turgor generation by the rice blast fungus Magnaporthe grisea". Biochemical Society Transactions. 33 (Pt 2): 384–8. doi:10.1042/BST0330384. PMID 15787612.
- Pereira JL, Noronha EF, Miller RN, Franco OL (June 2007). "Novel insights in the use of hydrolytic enzymes secreted by fungi with biotechnological potential". Letters in Applied Microbiology. 44 (6): 573–81. doi:10.1111/j.1472-765X.2007.02151.x. PMID 17576216.
- Schaller M, Borelli C, Korting HC, Hube B (November 2005). "Hydrolytic enzymes as virulence factors of Candida albicans". Mycoses. 48 (6): 365–77. doi:10.1111/j.1439-0507.2005.01165.x. PMID 16262871.
- Farrar JF (October 1985). "Carbohydrate metabolism in biotrophic plant pathogens". Microbiological Sciences. 2 (10): 314–7. PMID 3939987.
- Fischer R, Zekert N, Takeshita N (May 2008). "Polarized growth in fungi--interplay between the cytoskeleton, positional markers and membrane domains". Molecular Microbiology. 68 (4): 813–26. doi:10.1111/j.1365-2958.2008.06193.x. PMID 18399939.
- Christensen MJ, Bennett RJ, Ansari HA, Koga H, Johnson RD, Bryan GT, Simpson WR, Koolaard JP, Nickless EM, Voisey CR (February 2008). "Epichloë endophytes grow by intercalary hyphal extension in elongating grass leaves". Fungal Genetics and Biology. 45 (2): 84–93. doi:10.1016/j.fgb.2007.07.013. PMID 17919950.
- Money NP (October 2002). "Mushroom stem cells". BioEssays. 24 (10): 949–52. doi:10.1002/bies.10160. PMID 12325127.
- Willensdorfer M (February 2009). "On the evolution of differentiated multicellularity". Evolution; International Journal of Organic Evolution. 63 (2): 306–23. arXiv:0801.2610. doi:10.1111/j.1558-5646.2008.00541.x. PMID 19154376.
- Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR (May 2006). "Opaque cells signal white cells to form biofilms in Candida albicans". The EMBO Journal. 25 (10): 2240–52. doi:10.1038/sj.emboj.7601099. PMC 1462973. PMID 16628217.
- Marzluf GA (September 1981). "Regulation of nitrogen metabolism and gene expression in fungi". Microbiological Reviews. 45 (3): 437–61. PMC 281519. PMID 6117784.
- Hynes MJ (1994). "Regulatory circuits of the amdS gene of Aspergillus nidulans". Antonie van Leeuwenhoek. 65 (3): 179–82. doi:10.1007/BF00871944. PMID 7847883.
- Dadachova E, Casadevall A (December 2008). "Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin". Current Opinion in Microbiology. 11 (6): 525–31. doi:10.1016/j.mib.2008.09.013. PMC 2677413. PMID 18848901.
- Alexopoulos et al., pp. 48–56.
- Kirk et al., p. 633.
- Heitman J (September 2006). "Sexual reproduction and the evolution of microbial pathogens". Current Biology. 16 (17): R711–25. Bibcode:1996CBio....6.1213A. doi:10.1016/j.cub.2006.07.064. PMID 16950098.
- Alcamo IE, Pommerville J (2004). Alcamo's Fundamentals of Microbiology. Boston, Massachusetts: Jones and Bartlett. p. 590. ISBN 978-0-7637-0067-6.
- Redecker D, Raab P (2006). "Phylogeny of the glomeromycota (arbuscular mycorrhizal fungi): recent developments and new gene markers". Mycologia. 98 (6): 885–95. doi:10.3852/mycologia.98.6.885. PMID 17486965.
- Guarro J, Stchigel AM (July 1999). "Developments in fungal taxonomy". Clinical Microbiology Reviews. 12 (3): 454–500. doi:10.1128/CMR.12.3.454. PMC 100249. PMID 10398676.
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (October 2000). "Phylogenetic species recognition and species concepts in fungi". Fungal Genetics and Biology. 31 (1): 21–32. doi:10.1006/fgbi.2000.1228. PMID 11118132.
- Metzenberg RL, Glass NL (February 1990). "Mating type and mating strategies in Neurospora". BioEssays. 12 (2): 53–9. doi:10.1002/bies.950120202. PMID 2140508.
- Jennings and Lysek, pp. 107–114.
- Deacon, p. 31.
- Alexopoulos et al., pp. 492–493.
- Jennings and Lysek, p. 142.
- Deacon, pp. 21–24.
- Linder MB, Szilvay GR, Nakari-Setälä T, Penttilä ME (November 2005). "Hydrophobins: the protein-amphiphiles of filamentous fungi". FEMS Microbiology Reviews. 29 (5): 877–96. doi:10.1016/j.femsre.2005.01.004. PMID 16219510.
- Trail F (November 2007). "Fungal cannons: explosive spore discharge in the Ascomycota". FEMS Microbiology Letters. 276 (1): 12–8. doi:10.1111/j.1574-6968.2007.00900.x. PMID 17784861.
- Pringle A, Patek SN, Fischer M, Stolze J, Money NP (2005). "The captured launch of a ballistospore". Mycologia. 97 (4): 866–71. doi:10.3852/mycologia.97.4.866. PMID 16457355.
- Kirk et al., p. 495.
- Stipitate hydnoid fungi, Hampshire Biodiversity Partnership
- Brodie HJ (1975). The Bird's Nest Fungi. Toronto, Ontario: University of Toronto Press. p. 80. ISBN 978-0-8020-5307-7.
- Alexopoulos et al., p. 545.
- "Spore Dispersal in Fungi". www.botany.hawaii.edu. Retrieved 28 December 2018.
- "Dispersal". herbarium.usu.edu. Retrieved 28 December 2018.
- Dyer PS, O'Gorman CM. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev. 2012 Jan;36(1):165-92. doi:10.1111/j.1574-6976.2011.00308.x. Epub 2011 Oct 6. Review. PMID 22091779
- Yun SH, Berbee ML, Yoder OC, Turgeon BG. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci U S A. 1999 May 11;96(10):5592-7. PMID 10318929 PMCID: PMC21905 doi:10.1073/pnas.96.10.5592
- Richard S, Almeida JMGCF, Cissé OH, Luraschi A, Nielsen O, Pagni M, Hauser PM. Functional and Expression Analyses of the Pneumocystis MAT Genes Suggest Obligate Sexuality through Primary Homothallism within Host Lungs. MBio. 2018 Feb 20;9(1). pii: e02201-17. doi:10.1128/mBio.02201-17. PMID 29463658
- Heitman J. Evolution of sexual reproduction: a view from the Fungal Kingdom supports an evolutionary epoch with sex before sexes. Fungal Biol Rev. 2015 Dec 1;29(3-4):108-117. PMID 26834823
- Jennings and Lysek, pp. 114–115.
- Furlaneto MC, Pizzirani-Kleiner AA (January 1992). "Intraspecific hybridisation of Trichoderma pseudokoningii by anastomosis and by protoplast fusion". FEMS Microbiology Letters. 69 (2): 191–5. doi:10.1111/j.1574-6968.1992.tb05150.x. PMID 1537549.
- Schardl CL, Craven KD (November 2003). "Interspecific hybridization in plant-associated fungi and oomycetes: a review". Molecular Ecology. 12 (11): 2861–73. doi:10.1046/j.1365-294X.2003.01965.x. PMID 14629368.
- Donoghue MJ; Cracraft J (2004). Assembling the Tree of Life. Oxford (Oxfordshire), UK: Oxford University Press. p. 187. ISBN 978-0-19-517234-8.
- Taylor and Taylor, p. 19.
- Taylor and Taylor, pp. 7–12.
- Bengtson, Stefan; Rasmussen, Birger; Ivarsson, Magnus; Muhling, Janet; Broman, Curt; Marone, Federica; Stampanoni, Marco; Bekker, Andrey (24 April 2017). "Fungus-like mycelial fossils in 2.4-billion-year-old vesicular basalt". Nature Ecology & Evolution. 1 (6): 0141. doi:10.1038/s41559-017-0141. PMID 28812648.
- Lücking R, Huhndorf S, Pfister DH, Plata ER, Lumbsch HT (2009). "Fungi evolved right on track". Mycologia. 101 (6): 810–22. doi:10.3852/09-016. PMID 19927746.
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, et al. (October 2006). "Reconstructing the early evolution of Fungi using a six-gene phylogeny". Nature. 443 (7113): 818–22. Bibcode:2006Natur.443..818J. doi:10.1038/nature05110. PMID 17051209.
- Taylor and Taylor, pp. 84–94 and 106–107.
- Schoch CL, Sung GH, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, et al. (April 2009). "The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits". Systematic Biology. 58 (2): 224–39. doi:10.1093/sysbio/syp020. PMID 20525580.
- Zimmer, Carl (22 May 2019). "How Did Life Arrive on Land? A Billion-Year-Old Fungus May Hold Clues - A cache of microscopic fossils from the Arctic hints that fungi reached land long before plants". The New York Times. Retrieved 23 May 2019.
- Loron, Corentin C.; François, Camille; Rainbird, Robert H.; Turner, Elizabeth C.; Borensztajn, Stephan; Javaux, Emmanuelle J. (22 May 2019). "Early fungi from the Proterozoic era in Arctic Canada". Nature. Springer Science and Business Media LLC. 570 (7760): 232–235. Bibcode:2019Natur.570..232L. doi:10.1038/s41586-019-1217-0. ISSN 0028-0836. PMID 31118507.
- Timmer, John (22 May 2019). "Billion-year-old fossils may be early fungus". Ars Technica. Retrieved 23 May 2019.
- Brundrett MC (2002). "Coevolution of roots and mycorrhizas of land plants". New Phytologist. 154 (2): 275–304. doi:10.1046/j.1469-8137.2002.00397.x.
- Redecker D, Kodner R, Graham LE (September 2000). "Glomalean fungi from the Ordovician". Science. 289 (5486): 1920–1. Bibcode:2000Sci...289.1920R. doi:10.1126/science.289.5486.1920. PMID 10988069.
- Taylor TN, Taylor EL (1996). "The distribution and interactions of some Paleozoic fungi". Review of Palaeobotany and Palynology. 95 (1–4): 83–94. doi:10.1016/S0034-6667(96)00029-2.
- Dotzler N, Walker C, Krings M, Hass H, Kerp H, Taylor TN, Agerer R (2009). "Acaulosporoid glomeromycotan spores with a germination shield from the 400-million-year-old Rhynie chert" (PDF). Mycological Progress. 8 (1): 9–18. doi:10.1007/s11557-008-0573-1. hdl:1808/13680.
- Taylor JW, Berbee ML (2006). "Dating divergences in the Fungal Tree of Life: review and new analyses". Mycologia. 98 (6): 838–49. doi:10.3852/mycologia.98.6.838. PMID 17486961.
- Blackwell M, Vilgalys R, James TY, Taylor JW (2009). "Fungi. Eumycota: mushrooms, sac fungi, yeast, molds, rusts, smuts, etc". Tree of Life Web Project. Retrieved 25 April 2009.
- Yuan X, Xiao S, Taylor TN (May 2005). "Lichen-like symbiosis 600 million years ago". Science. 308 (5724): 1017–20. Bibcode:2005Sci...308.1017Y. doi:10.1126/science.1111347. PMID 15890881.
- Karatygin IV, Snigirevskaya NS, Vikulin SV (2009). "The most ancient terrestrial lichen Winfrenatia reticulata: A new find and new interpretation". Paleontological Journal. 43 (1): 107–114. doi:10.1134/S0031030109010110.
- Taylor TN, Hass H, Kerp H, Krings M, Hanlin RT (2005). "Perithecial ascomycetes from the 400 million year old Rhynie chert: an example of ancestral polymorphism". Mycologia. 97 (1): 269–85. doi:10.3852/mycologia.97.1.269. hdl:1808/16786. PMID 16389979.
- Dennis RL (1970). "A Middle Pennsylvanian basidiomycete mycelium with clamp connections". Mycologia. 62 (3): 578–584. doi:10.2307/3757529. JSTOR 3757529.
- Hibbett DS, Grimaldi D, Donoghue MJ (1995). "Cretaceous mushrooms in amber". Nature. 377 (6549): 487. Bibcode:1995Natur.377..487H. doi:10.1038/377487a0.
- Hibbett DS, Grimaldi D, Donoghue MJ (1997). "Fossil mushrooms from Miocene and Cretaceous ambers and the evolution of homobasidiomycetes". American Journal of Botany. 84 (7): 981–991. doi:10.2307/2446289. JSTOR 2446289.
- Eshet Y, Rampino MR, Visscher H (1995). "Fungal event and palynological record of ecological crisis and recovery across the Permian-Triassic boundary". Geology. 23 (1): 967–970. Bibcode:1995Geo....23..967E. doi:10.1130/0091-7613(1995)023<0967:FEAPRO>2.3.CO;2.
- Foster CB, Stephenson MH, Marshall C, Logan GA, Greenwood PF (2002). "A revision of Reduviasporonites Wilson 1962: description, illustration, comparison and biological affinities". Palynology. 26 (1): 35–58. doi:10.2113/0260035.
- López-Gómez J, Taylor EL (2005). "Permian-Triassic transition in Spain: a multidisciplinary approach". Palaeogeography, Palaeoclimatology, Palaeoecology. 229 (1–2): 1–2. doi:10.1016/j.palaeo.2005.06.028.
- Looy CV, Twitchett RJ, Dilcher DL, Van Konijnenburg-Van Cittert JH, Visscher H (July 2001). "Life in the end-Permian dead zone". Proceedings of the National Academy of Sciences of the United States of America. 98 (14): 7879–83. Bibcode:2001PNAS...98.7879L. doi:10.1073/pnas.131218098. PMC 35436. PMID 11427710.
See image 2
- Ward PD, Botha J, Buick R, De Kock MO, Erwin DH, Garrison GH, Kirschvink JL, Smith R (February 2005). "Abrupt and gradual extinction among Late Permian land vertebrates in the Karoo basin, South Africa". Science. 307 (5710): 709–14. Bibcode:2005Sci...307..709W. CiteSeerX 10.1.1.503.2065. doi:10.1126/science.1107068. PMID 15661973.
- Casadevall, Arturo; Heitman, Joseph (16 August 2012). "Fungi and the Rise of Mammals". PLoS Pathogens. 8 (8): e1002808. doi:10.1371/journal.ppat.1002808. PMC 3420938. PMID 22916007.
That ecological calamity was accompanied by massive deforestation, an event followed by a fungal bloom, as the earth became a massive compost.
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T (2008). "Multigene phylogeny of choanozoa and the origin of animals". PLOS ONE. 3 (5): e2098. Bibcode:2008PLoSO...3.2098S. doi:10.1371/journal.pone.0002098. PMC 2346548. PMID 18461162.

- See Palaeos Fungi: Fungi for an introduction to fungal taxonomy, including recent controversies. archive
- Celio GJ, Padamsee M, Dentinger BT, Bauer R, McLaughlin DJ (2006). "Assembling the Fungal Tree of Life: constructing the structural and biochemical database". Mycologia. 98 (6): 850–9. doi:10.3852/mycologia.98.6.850. PMID 17486962.
- Silar P (2016). Protistes Eucaryotes: Origine, Evolution et Biologie des Microbes Eucaryotes. HAL. p. 462. ISBN 978-2-9555841-0-1.
- Esser K (2014). The Mycota VII A: Systematics and Evolution (2nd ed.). Springer. p. 461. ISBN 978-3-642-55317-2.
- Tedersoo, Leho; Sanchez-Ramırez, Santiago; Koljalg, Urmas; Bahram, Mohammad; Doring, Markus; Schigel, Dmitry; May, Tom; Ryberg, Martin; Abarenkov, Kessy (22 February 2018). "High-level classification of the Fungi and a tool for evolutionary ecological analyses". Fungal Diversity. 90 (1): 135–159. doi:10.1007/s13225-018-0401-0.
- Gill EE, Fast NM (June 2006). "Assessing the microsporidia-fungi relationship: Combined phylogenetic analysis of eight genes". Gene. 375: 103–9. doi:10.1016/j.gene.2006.02.023. PMID 16626896.
- Liu YJ, Hodson MC, Hall BD (2006). "Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes". BMC Evolutionary Biology. 6: 74. doi:10.1186/1471-2148-6-74. PMC 1599754. PMID 17010206.
- James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, Powell MJ, Griffith GW, Vilgalys R (2006). "A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota)". Mycologia. 98 (6): 860–71. doi:10.3852/mycologia.98.6.860. PMID 17486963.
- Lockhart RJ, Van Dyke MI, Beadle IR, Humphreys P, McCarthy AJ (August 2006). "Molecular biological detection of anaerobic gut fungi (Neocallimastigales) from landfill sites". Applied and Environmental Microbiology. 72 (8): 5659–61. doi:10.1128/AEM.01057-06. PMC 1538735. PMID 16885325.
- Remy W, Taylor TN, Hass H, Kerp H (December 1994). "Four hundred-million-year-old vesicular arbuscular mycorrhizae". Proceedings of the National Academy of Sciences of the United States of America. 91 (25): 11841–3. Bibcode:1994PNAS...9111841R. doi:10.1073/pnas.91.25.11841. PMC 45331. PMID 11607500.
- Schüssler A, Schwarzott D, Walker C (2001). "A new fungal phylum, the Glomeromycota: phylogeny and evolution". Mycological Research. 105 (12): 1413–1421. doi:10.1017/S0953756201005196.
- Alexopoulos et al., p. 145.
- For an example, see Samuels GJ (February 2006). "Trichoderma: systematics, the sexual state, and ecology". Phytopathology. 96 (2): 195–206. doi:10.1094/PHYTO-96-0195. PMID 18943925.
- Radford A, Parish JH (June 1997). "The genome and genes of Neurospora crassa". Fungal Genetics and Biology. 21 (3): 258–66. doi:10.1006/fgbi.1997.0979. PMID 9290240.
- Valverde ME, Paredes-López O, Pataky JK, Guevara-Lara F (January 1995). "Huitlacoche (Ustilago maydis) as a food source--biology, composition, and production". Critical Reviews in Food Science and Nutrition. 35 (3): 191–229. doi:10.1080/10408399509527699. PMID 7632354.
- Zisova LG (2009). "Malassezia species and seborrheic dermatitis". Folia Medica. 51 (1): 23–33. PMID 19437895.
- Perfect JR (June 2006). "Cryptococcus neoformans: the yeast that likes it hot". FEMS Yeast Research. 6 (4): 463–8. doi:10.1111/j.1567-1364.2006.00051.x. PMID 16696642.
- Blackwell M, Spatafora JW (2004). "Fungi and their allies". In Bills GF, Mueller GM, Foster MS (eds.). Biodiversity of Fungi: Inventory and Monitoring Methods. Amsterdam: Elsevier Academic Press. pp. 18–20. ISBN 978-0-12-509551-8.
- Amoroso, Maria Julia; Benimeli, Claudia Susana; Cuozzo, Sergio Antonio (2013). Actinobacteria : application in bioremediation and production of industrial enzymes. CRC Press, Taylor & Francis Group. p. 33. ISBN 9781466578739.
- An introduction to soil biology, Actinobacteria
- Gadd GM (January 2007). "Geomycology: biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation". Mycological Research. 111 (Pt 1): 3–49. doi:10.1016/j.mycres.2006.12.001. PMID 17307120.
- Lindahl BD, Ihrmark K, Boberg J, Trumbore SE, Högberg P, Stenlid J, Finlay RD (2007). "Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest". The New Phytologist. 173 (3): 611–20. doi:10.1111/j.1469-8137.2006.01936.x. PMID 17244056.
- Barea JM, Pozo MJ, Azcón R, Azcón-Aguilar C (July 2005). "Microbial co-operation in the rhizosphere". Journal of Experimental Botany. 56 (417): 1761–78. doi:10.1093/jxb/eri197. PMID 15911555.
- Aanen DK (June 2006). "As you reap, so shall you sow: coupling of harvesting and inoculating stabilizes the mutualism between termites and fungi". Biology Letters. 2 (2): 209–12. doi:10.1098/rsbl.2005.0424. PMC 1618886. PMID 17148364.
- Nikoh N, Fukatsu T (April 2000). "Interkingdom host jumping underground: phylogenetic analysis of entomoparasitic fungi of the genus cordyceps". Molecular Biology and Evolution. 17 (4): 629–38. doi:10.1093/oxfordjournals.molbev.a026341. PMID 10742053.
- Perotto S, Bonfante P (December 1997). "Bacterial associations with mycorrhizal fungi: close and distant friends in the rhizosphere". Trends in Microbiology. 5 (12): 496–501. doi:10.1016/S0966-842X(97)01154-2. PMID 9447662.
- Arnold AE, Mejía LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA (December 2003). "Fungal endophytes limit pathogen damage in a tropical tree". Proceedings of the National Academy of Sciences of the United States of America. 100 (26): 15649–54. Bibcode:2003PNAS..10015649A. doi:10.1073/pnas.2533483100. PMC 307622. PMID 14671327.
- Paszkowski U (August 2006). "Mutualism and parasitism: the yin and yang of plant symbioses". Current Opinion in Plant Biology. 9 (4): 364–70. doi:10.1016/j.pbi.2006.05.008. PMID 16713732.
- Hube B (August 2004). "From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans". Current Opinion in Microbiology. 7 (4): 336–41. doi:10.1016/j.mib.2004.06.003. PMID 15288621.
- Bonfante P (April 2003). "Plants, mycorrhizal fungi and endobacteria: a dialog among cells and genomes". The Biological Bulletin. 204 (2): 215–20. doi:10.2307/1543562. JSTOR 1543562. PMID 12700157.
- van der Heijden MG, Streitwolf-Engel R, Riedl R, Siegrist S, Neudecker A, Ineichen K, Boller T, Wiemken A, Sanders IR (2006). "The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland". The New Phytologist. 172 (4): 739–52. doi:10.1111/j.1469-8137.2006.01862.x. PMID 17096799.
- Heijden, Marcel G. A. van der (15 April 2016). "Underground networking". Science. 352 (6283): 290–291. Bibcode:2016Sci...352..290H. doi:10.1126/science.aaf4694. ISSN 0036-8075. PMID 27081054.
- Selosse MA, Richard F, He X, Simard SW (November 2006). "Mycorrhizal networks: des liaisons dangereuses?". Trends in Ecology & Evolution. 21 (11): 621–8. doi:10.1016/j.tree.2006.07.003. PMID 16843567.
- Yong, Ed (14 April 2016). "Trees Have Their Own Internet". The Atlantic. Retrieved 9 March 2019.
- Merckx V, Bidartondo MI, Hynson NA (December 2009). "Myco-heterotrophy: when fungi host plants". Annals of Botany. 104 (7): 1255–61. doi:10.1093/aob/mcp235. PMC 2778383. PMID 19767309.
- Schulz B, Boyle C (June 2005). "The endophytic continuum". Mycological Research. 109 (Pt 6): 661–86. doi:10.1017/S095375620500273X. PMID 16080390.
- Clay K, Schardl C (October 2002). "Evolutionary origins and ecological consequences of endophyte symbiosis with grasses". The American Naturalist. 160 Suppl 4 (suppl. 4): S99–S127. doi:10.1086/342161. PMID 18707456.
- Brodo IM, Sharnoff SD (2001). Lichens of North America. New Haven, Connecticut: Yale University Press. ISBN 978-0-300-08249-4.
- Raven PH, Evert RF, Eichhorn, SE (2005). "14—Fungi". Biology of Plants (7 ed.). W. H. Freeman. p. 290. ISBN 978-0-7167-1007-3.
- Deacon, p. 267.
- Purvis W (2000). Lichens. Washington, D.C.: Smithsonian Institution Press in association with the Natural History Museum, London. pp. 49–75. ISBN 978-1-56098-879-3.
- Kirk et al., p. 378.
- Deacon, pp. 267–276.
- Douglas AE (November 1989). "Mycetocyte symbiosis in insects". Biological Reviews of the Cambridge Philosophical Society. 64 (4): 409–34. doi:10.1111/j.1469-185X.1989.tb00682.x. PMID 2696562.
- Deacon, p. 277.
- "Entomologists: Brazilian Stingless Bee Must Cultivate Special Type of Fungus to Survive". Sci-News.com. 23 October 2015. Retrieved 25 October 2015.
- Nguyen NH, Suh SO, Blackwell M (2007). "Five novel Candida species in insect-associated yeast clades isolated from Neuroptera and other insects". Mycologia. 99 (6): 842–58. doi:10.3852/mycologia.99.6.842. PMID 18333508.
- Filipiak, Michał; Weiner, January (March 2017). "Nutritional dynamics during the development of xylophagous beetles related to changes in the stoichiometry of 11 elements". Physiological Entomology. 42 (1): 73–84. doi:10.1111/phen.12168.
- Ulyshen, Michael D. (2018). Saproxylic insects : diversity, ecology and conservation. Springer, Cham. p. 429–469. doi:10.1007/978-3-319-75937-1_13. ISBN 978-3-319-75937-1.
- Ulyshen, Michael D. (2018). Saproxylic Insects Diversity, Ecology and Conservation. Springer, Cham. p. 377–427. doi:10.1007/978-3-319-75937-1_12. ISBN 978-3-319-75936-4.
- Filipiak, Michał; Sobczyk, Łukasz; Weiner, January (2016). "Fungal Transformation of Tree Stumps into a Suitable Resource for Xylophagous Beetles via Changes in Elemental Ratios". Insects. 7 (2): 13. doi:10.3390/insects7020013. PMC 4931425.
- Chandler PJ (2010). A Dipterist's Handbook (2nd Edition). U.K.: The Amateur Entomologists' Society. pp. 1–525.
- Talbot NJ (2003). "On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea". Annual Review of Microbiology. 57: 177–202. doi:10.1146/annurev.micro.57.030502.090957. PMID 14527276.
- Paoletti M, Buck KW, Brasier CM (January 2006). "Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi". Molecular Ecology. 15 (1): 249–62. doi:10.1111/j.1365-294X.2005.02728.x. PMID 16367844.
- Gryzenhout M, Wingfield BD, Wingfield MJ (May 2006). "New taxonomic concepts for the important forest pathogen Cryphonectria parasitica and related fungi". FEMS Microbiology Letters. 258 (2): 161–72. doi:10.1111/j.1574-6968.2006.00170.x. PMID 16640568.
- Yang Y, Yang E, An Z, Liu X (May 2007). "Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences". Proceedings of the National Academy of Sciences of the United States of America. 104 (20): 8379–84. Bibcode:2007PNAS..104.8379Y. doi:10.1073/pnas.0702770104. PMC 1895958. PMID 17494736.
- Koeck, M.; Hardham, A.R.; Dodds; P.N. (2011). "The role of effectors of biotrophic and hemibiotrophic fungi in infection". Cellular Microbiology. 13 (12): 1849–1857. doi:10.1111/j.1462-5822.2011.01665.x. PMC 3218205. PMID 21848815.
- "Asterotremella gen. nov. albida, an anamorphic tremelloid yeast isolated from the agarics Asterophora lycoperdoides and Asterophora parasitica". ResearchGate. Retrieved 19 April 2019.
- Nielsen K, Heitman J (2007). "Sex and Virulence of Human Pathogenic Fungi". Fungal Genomics. Advances in Genetics. 57. pp. 143–73. doi:10.1016/S0065-2660(06)57004-X. ISBN 978-0-12-017657-1. PMID 17352904.
- Brakhage AA (December 2005). "Systemic fungal infections caused by Aspergillus species: epidemiology, infection process and virulence determinants". Current Drug Targets. 6 (8): 875–86. doi:10.2174/138945005774912717. PMID 16375671.
- Kauffman CA (January 2007). "Histoplasmosis: a clinical and laboratory update". Clinical Microbiology Reviews. 20 (1): 115–32. doi:10.1128/CMR.00027-06. PMC 1797635. PMID 17223625.
- Cushion MT, Smulian AG, Slaven BE, Sesterhenn T, Arnold J, Staben C, Porollo A, Adamczak R, Meller J (2007). "Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite". PLOS ONE. 2 (5): e423. Bibcode:2007PLoSO...2..423C. doi:10.1371/journal.pone.0000423. PMC 1855432. PMID 17487271.

- Cook GC, Zumla AI (2008). Manson's Tropical Diseases: Expert Consult. Edinburgh, Scotland: Saunders Ltd. p. 347. ISBN 978-1-4160-4470-3.
- Simon-Nobbe B, Denk U, Pöll V, Rid R, Breitenbach M (2008). "The spectrum of fungal allergy". International Archives of Allergy and Immunology. 145 (1): 58–86. doi:10.1159/000107578. PMID 17709917.
- Le Floch G, Rey P, Benizri E, Benhamou N, Tirilly Y (2003). "Impact of auxin-compounds produced by the antagonistic fungus Pythium oligandrum or the minor pathogen Pythium group F on plant growth". Plant Soil. 257 (2): 459–470. doi:10.1023/a:1027330024834.
- Pearson MN, Beever RE, Boine B, Arthur K (January 2009). "Mycoviruses of filamentous fungi and their relevance to plant pathology". Molecular Plant Pathology. 10 (1): 115–28. doi:10.1111/j.1364-3703.2008.00503.x. PMC 6640375. PMID 19161358.
- Bozarth RF (October 1972). "Mycoviruses: a new dimension in microbiology". Environmental Health Perspectives. 2 (1): 23–39. doi:10.1289/ehp.720223. PMC 1474899. PMID 4628853.
- Schardl CL, Panaccione DG, Tudzynski P (2006). Ergot alkaloids--biology and molecular biology. The Alkaloids: Chemistry and Biology. 63. pp. 45–86. doi:10.1016/S1099-4831(06)63002-2. ISBN 978-0-12-469563-4. PMID 17133714.
- van Egmond HP, Schothorst RC, Jonker MA (September 2007). "Regulations relating to mycotoxins in food: perspectives in a global and European context". Analytical and Bioanalytical Chemistry. 389 (1): 147–57. doi:10.1007/s00216-007-1317-9. PMID 17508207.
- Demain AL, Fang A (2000). "The Natural Functions of Secondary Metabolites". History of Modern Biotechnology I. Advances in Biochemical Engineering/Biotechnology. 69. pp. 1–39. doi:10.1007/3-540-44964-7_1. ISBN 978-3-540-67793-2. PMID 11036689.
- Rohlfs M, Albert M, Keller NP, Kempken F (October 2007). "Secondary chemicals protect mould from fungivory". Biology Letters. 3 (5): 523–5. doi:10.1098/rsbl.2007.0338. PMC 2391202. PMID 17686752.
- Molina L, Kahmann R (July 2007). "An Ustilago maydis gene involved in H2O2 detoxification is required for virulence". The Plant Cell. 19 (7): 2293–309. doi:10.1105/tpc.107.052332. PMC 1955693. PMID 17616735.
- Kojic M, Zhou Q, Lisby M, Holloman WK (January 2006). "Rec2 interplay with both Brh2 and Rad51 balances recombinational repair in Ustilago maydis". Molecular and Cellular Biology. 26 (2): 678–88. doi:10.1128/MCB.26.2.678-688.2006. PMC 1346908. PMID 16382157.
- Michod RE, Bernstein H, Nedelcu AM (May 2008). "Adaptive value of sex in microbial pathogens" (PDF). Infection, Genetics and Evolution. 8 (3): 267–85. doi:10.1016/j.meegid.2008.01.002. PMID 18295550.
- Fan W, Kraus PR, Boily MJ, Heitman J (August 2005). "Cryptococcus neoformans gene expression during murine macrophage infection". Eukaryotic Cell. 4 (8): 1420–33. doi:10.1128/EC.4.8.1420-1433.2005. PMC 1214536. PMID 16087747.
- Lin X, Hull CM, Heitman J (April 2005). "Sexual reproduction between partners of the same mating type in Cryptococcus neoformans". Nature. 434 (7036): 1017–21. Bibcode:2005Natur.434.1017L. doi:10.1038/nature03448. PMID 15846346.
- Fincham JR (March 1989). "Transformation in fungi". Microbiological Reviews. 53 (1): 148–70. PMC 372721. PMID 2651864.
- Hawkins KM, Smolke CD (September 2008). "Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae". Nature Chemical Biology. 4 (9): 564–73. doi:10.1038/nchembio.105. PMC 2830865. PMID 18690217.
- Huang B, Guo J, Yi B, Yu X, Sun L, Chen W (July 2008). "Heterologous production of secondary metabolites as pharmaceuticals in Saccharomyces cerevisiae". Biotechnology Letters. 30 (7): 1121–37. doi:10.1007/s10529-008-9663-z. PMID 18512022.
- Brakhage AA, Spröte P, Al-Abdallah Q, Gehrke A, Plattner H, Tüncher A (2004). "Regulation of Penicillin Biosynthesis in Filamentous Fungi". Molecular Biotechnology of Fungal beta-Lactam Antibiotics and Related Peptide Synthetases. Advances in Biochemical Engineering/Biotechnology. 88. pp. 45–90. doi:10.1007/b99257. ISBN 978-3-540-22032-9. PMID 15719552.
- Pan A, Lorenzotti S, Zoncada A (January 2008). "Registered and investigational drugs for the treatment of methicillin-resistant Staphylococcus aureus infection". Recent Patents on Anti-Infective Drug Discovery. 3 (1): 10–33. doi:10.2174/157489108783413173. PMID 18221183.
- Fajardo A, Martínez JL (April 2008). "Antibiotics as signals that trigger specific bacterial responses". Current Opinion in Microbiology. 11 (2): 161–7. doi:10.1016/j.mib.2008.02.006. PMID 18373943.
- Loo DS (2006). "Systemic antifungal agents: an update of established and new therapies". Advances in Dermatology. 22: 101–24. doi:10.1016/j.yadr.2006.07.001. PMID 17249298.
- Manzoni M, Rollini M (April 2002). "Biosynthesis and biotechnological production of statins by filamentous fungi and application of these cholesterol-lowering drugs". Applied Microbiology and Biotechnology. 58 (5): 555–64. doi:10.1007/s00253-002-0932-9. PMID 11956737.
- el-Mekkawy S, Meselhy MR, Nakamura N, Tezuka Y, Hattori M, Kakiuchi N, Shimotohno K, Kawahata T, Otake T (November 1998). "Anti-HIV-1 and anti-HIV-1-protease substances from Ganoderma lucidum". Phytochemistry. 49 (6): 1651–7. doi:10.1016/S0031-9422(98)00254-4. PMID 9862140.
- El Dine RS, El Halawany AM, Ma CM, Hattori M (June 2008). "Anti-HIV-1 protease activity of lanostane triterpenes from the vietnamese mushroom Ganoderma colossum". Journal of Natural Products. 71 (6): 1022–6. doi:10.1021/np8001139. PMID 18547117.
- Hetland G, Johnson E, Lyberg T, Bernardshaw S, Tryggestad AM, Grinde B (October 2008). "Effects of the medicinal mushroom Agaricus blazei Murill on immunity, infection and cancer". Scandinavian Journal of Immunology. 68 (4): 363–70. doi:10.1111/j.1365-3083.2008.02156.x. PMID 18782264.
- Yuen JW, Gohel MD (2005). "Anticancer effects of Ganoderma lucidum: a review of scientific evidence". Nutrition and Cancer. 53 (1): 11–7. doi:10.1207/s15327914nc5301_2. hdl:10397/26313. PMID 16351502.
- Sullivan R, Smith JE, Rowan NJ (2006). "Medicinal mushrooms and cancer therapy: translating a traditional practice into Western medicine". Perspectives in Biology and Medicine. 49 (2): 159–70. doi:10.1353/pbm.2006.0034. PMID 16702701.
- Halpern GM, Miller A (2002). Medicinal Mushrooms: Ancient Remedies for Modern Ailments. New York, New York: M. Evans and Co. p. 116. ISBN 978-0-87131-981-4.
- Fisher M, Yang LX (2002). "Anticancer effects and mechanisms of polysaccharide-K (PSK): implications of cancer immunotherapy". Anticancer Research. 22 (3): 1737–54. PMID 12168863.
- Firenzuoli F, Gori L, Lombardo G (March 2008). "The Medicinal Mushroom Agaricus blazei Murrill: Review of Literature and Pharmaco-Toxicological Problems". Evidence-Based Complementary and Alternative Medicine. 5 (1): 3–15. doi:10.1093/ecam/nem007. PMC 2249742. PMID 18317543.
- Paterson RR (September 2006). "Ganoderma - a therapeutic fungal biofactory". Phytochemistry. 67 (18): 1985–2001. doi:10.1016/j.phytochem.2006.07.004. hdl:1822/5522. PMID 16905165.
- Paterson RR (May 2008). "Cordyceps: a traditional Chinese medicine and another fungal therapeutic biofactory?". Phytochemistry. 69 (7): 1469–95. doi:10.1016/j.phytochem.2008.01.027. hdl:1822/7896. PMID 18343466.
- Kulp K (2000). Handbook of Cereal Science and Technology. CRC Press. ISBN 978-0-8247-8294-8.
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C (April 2006). "How did Saccharomyces evolve to become a good brewer?". Trends in Genetics. 22 (4): 183–6. doi:10.1016/j.tig.2006.02.002. PMID 16499989.
- Abe K, Gomi K, Hasegawa F, Machida M (September 2006). "Impact of Aspergillus oryzae genomics on industrial production of metabolites". Mycopathologia. 162 (3): 143–53. doi:10.1007/s11046-006-0049-2. PMID 16944282.
- Hachmeister KA, Fung DY (1993). "Tempeh: a mold-modified indigenous fermented food made from soybeans and/or cereal grains". Critical Reviews in Microbiology. 19 (3): 137–88. doi:10.3109/10408419309113527. PMID 8267862.
- Jørgensen TR (December 2007). "Identification and toxigenic potential of the industrially important fungi, Aspergillus oryzae and Aspergillus sojae". Journal of Food Protection. 70 (12): 2916–34. doi:10.4315/0362-028X-70.12.2916. PMID 18095455.
- O'Donnell K, Cigelnik E, Casper HH (February 1998). "Molecular phylogenetic, morphological, and mycotoxin data support reidentification of the Quorn mycoprotein fungus as Fusarium venenatum". Fungal Genetics and Biology. 23 (1): 57–67. doi:10.1006/fgbi.1997.1018. PMID 9501477.
- Stamets P (2000). Growing Gourmet and Medicinal Mushrooms=[Shokuyō oyobi yakuyō kinoko no saibai]. Berkeley, California: Ten Speed Press. pp. 233–248. ISBN 978-1-58008-175-7.
- Hall, pp. 13–26.
- Kinsella JE, Hwang DH (November 1976). "Enzymes of Penicillium roqueforti involved in the biosynthesis of cheese flavor". Critical Reviews in Food Science and Nutrition. 8 (2): 191–228. doi:10.1080/10408397609527222. PMID 21770.
- Erdogan A, Gurses M, Sert S (August 2003). "Isolation of moulds capable of producing mycotoxins from blue mouldy Tulum cheeses produced in Turkey". International Journal of Food Microbiology. 85 (1–2): 83–5. doi:10.1016/S0168-1605(02)00485-3. PMID 12810273.
- Orr DB, Orr RT (1979). Mushrooms of Western North America. Berkeley, California: University of California Press. p. 17. ISBN 978-0-520-03656-7.
- Vetter J (January 1998). "Toxins of Amanita phalloides". Toxicon. 36 (1): 13–24. doi:10.1016/S0041-0101(97)00074-3. PMID 9604278.
- Leathem AM, Dorran TJ (March 2007). "Poisoning due to raw Gyromitra esculenta (false morels) west of the Rockies". Canadian Journal of Emergency Medicine. 9 (2): 127–30. doi:10.1017/s1481803500014937. PMID 17391587.
- Karlson-Stiber C, Persson H (September 2003). "Cytotoxic fungi--an overview". Toxicon. 42 (4): 339–49. doi:10.1016/S0041-0101(03)00238-1. PMID 14505933.
- Michelot D, Melendez-Howell LM (February 2003). "Amanita muscaria: chemistry, biology, toxicology, and ethnomycology". Mycological Research. 107 (Pt 2): 131–46. doi:10.1017/S0953756203007305. PMID 12747324.
- Hall, p. 7.
- Ammirati JF, McKenny M, Stuntz DE (1987). The New Savory Wild Mushroom. Seattle, Washington: University of Washington Press. pp. xii–xiii. ISBN 978-0-295-96480-5.
- López-Gómez J, Molina-Meyer M (February 2006). "The competitive exclusion principle versus biodiversity through competitive segregation and further adaptation to spatial heterogeneities". Theoretical Population Biology. 69 (1): 94–109. doi:10.1016/j.tpb.2005.08.004. PMID 16223517.
- Becker H (1998). "Setting the Stage To Screen Biocontrol Fungi". United States Department of Agriculture, Agricultural Research Service. Retrieved 23 February 2009.
- Keiller TS. "Whey-based fungal microfactory technology for enhanced biological pest management using fungi" (PDF). UVM Innovations. Archived from the original (PDF) on 29 October 2011. Retrieved 29 October 2011.
- Deshpande MV (1999). "Mycopesticide production by fermentation: potential and challenges". Critical Reviews in Microbiology. 25 (3): 229–43. doi:10.1080/10408419991299220. PMID 10524330.
- Thomas MB, Read AF (May 2007). "Can fungal biopesticides control malaria?". Nature Reviews. Microbiology. 5 (5): 377–83. doi:10.1038/nrmicro1638. PMID 17426726.
- Bush LP, Wilkinson HH, Schardl CL (May 1997). "Bioprotective Alkaloids of Grass-Fungal Endophyte Symbioses". Plant Physiology. 114 (1): 1–7. doi:10.1104/pp.114.1.1. PMC 158272. PMID 12223685.
- Bouton JH, Latch GC, Hill NS, Hoveland CS, McCann MA, Watson RH, Parish JA, Hawkins LL, Thompson FN (2002). "Reinfection of Tall Fescue Cultivars with Non-Ergot Alkaloid–Producing Endophytes". Agronomy Journal. 94 (3): 567–574. doi:10.2134/agronj2002.5670.
- Parish JA, McCann MA, Watson RH, Hoveland CS, Hawkins LL, Hill NS, Bouton JH (May 2003). "Use of nonergot alkaloid-producing endophytes for alleviating tall fescue toxicosis in sheep". Journal of Animal Science. 81 (5): 1316–22. doi:10.2527/2003.8151316x. PMID 12772860.
- Christian V, Shrivastava R, Shukla D, Modi HA, Vyas BR (April 2005). "Degradation of xenobiotic compounds by lignin-degrading white-rot fungi: enzymology and mechanisms involved". Indian Journal of Experimental Biology. 43 (4): 301–12. PMID 15875713.
- "Fungi to fight 'toxic war zones'". BBC News. 5 May 2008.
- Fomina M, Charnock JM, Hillier S, Alvarez R, Gadd GM (July 2007). "Fungal transformations of uranium oxides". Environmental Microbiology. 9 (7): 1696–710. doi:10.1111/j.1462-2920.2007.01288.x. PMID 17564604.
- Fomina M, Charnock JM, Hillier S, Alvarez R, Livens F, Gadd GM (May 2008). "Role of fungi in the biogeochemical fate of depleted uranium". Current Biology. 18 (9): R375–7. Bibcode:1996CBio....6.1213A. doi:10.1016/j.cub.2008.03.011. PMID 18460315.
- Beadle GW, Tatum EL (November 1941). "Genetic Control of Biochemical Reactions in Neurospora". Proceedings of the National Academy of Sciences of the United States of America. 27 (11): 499–506. Bibcode:1941PNAS...27..499B. doi:10.1073/pnas.27.11.499. PMC 1078370. PMID 16588492.
- Datta A, Ganesan K, Natarajan K (1989). "Current Trends in Candida albicans Research". Current trends in Candida albicans research. Advances in Microbial Physiology. 30. pp. 53–88. doi:10.1016/S0065-2911(08)60110-1. ISBN 978-0-12-027730-8. PMID 2700541.
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, et al. (April 2005). "The genome sequence of the rice blast fungus Magnaporthe grisea". Nature. 434 (7036): 980–6. Bibcode:2005Natur.434..980D. doi:10.1038/nature03449. PMID 15846337.
- Daly R, Hearn MT (2005). "Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production". Journal of Molecular Recognition. 18 (2): 119–38. doi:10.1002/jmr.687. PMID 15565717.
- Schlegel HG (1993). General Microbiology. Cambridge, UK: Cambridge University Press. p. 360. ISBN 978-0-521-43980-0.
- Joseph B, Ramteke PW, Thomas G (2008). "Cold active microbial lipases: some hot issues and recent developments". Biotechnology Advances. 26 (5): 457–70. doi:10.1016/j.biotechadv.2008.05.003. PMID 18571355.
- Kumar R, Singh S, Singh OV (May 2008). "Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives". Journal of Industrial Microbiology & Biotechnology. 35 (5): 377–91. doi:10.1007/s10295-008-0327-8. PMID 18338189.
- "Trichoderma spp., including T. harzianum, T. viride, T. koningii, T. hamatum and other spp. Deuteromycetes, Moniliales (asexual classification system)". Biological Control: A Guide to Natural Enemies in North America. Archived from the original on 14 April 2011. Retrieved 10 July 2007.
- Olempska-Beer ZS, Merker RI, Ditto MD, DiNovi MJ (July 2006). "Food-processing enzymes from recombinant microorganisms--a review". Regulatory Toxicology and Pharmacology. 45 (2): 144–158. doi:10.1016/j.yrtph.2006.05.001. PMID 16769167.
- Polizeli ML, Rizzatti AC, Monti R, Terenzi HF, Jorge JA, Amorim DS (June 2005). "Xylanases from fungi: properties and industrial applications". Applied Microbiology and Biotechnology. 67 (5): 577–91. doi:10.1007/s00253-005-1904-7. PMID 15944805.
Cited literature
- Ainsworth GC (1976). Introduction to the History of Mycology. Cambridge, UK: Cambridge University Press. ISBN 978-0-521-11295-6.
- Alexopoulos CJ, Mims CW, Blackwell M (1996). Introductory Mycology. John Wiley and Sons. ISBN 978-0-471-52229-4.
- Chandler PJ (2010). A Dipterist's Handbook (2nd ed.). The Amateur Entomologists' Society. pp. 1–525.
- Deacon J (2005). Fungal Biology. Cambridge, Massachusetts: Blackwell Publishers. ISBN 978-1-4051-3066-0.
- Hall IR (2003). Edible and Poisonous Mushrooms of the World. Portland, Oregon: Timber Press. ISBN 978-0-88192-586-9.
- Hanson JR (2008). The Chemistry of Fungi. Royal Society Of Chemistry. ISBN 978-0-85404-136-7.
- Jennings DH, Lysek G (1996). Fungal Biology: Understanding the Fungal Lifestyle. Guildford, UK: Bios Scientific Publishers Ltd. ISBN 978-1-85996-150-6.
- Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008). Dictionary of the Fungi (10th ed.). Wallingford, UK: CAB International. ISBN 978-0-85199-826-8.
- Taylor EL, Taylor TN (1993). The Biology and Evolution of Fossil Plants. Englewood Cliffs, New Jersey: Prentice Hall. ISBN 978-0-13-651589-0.
External links
| Look up fungus in Wiktionary, the free dictionary. |
| Library resources about Fungus |
- Tree of Life web project: Fungi
- Mushroom Observer (mushroomobserver.org), a collaborative fungus recording and identification project
- FUNGI in BoDD – Botanical Dermatology Database