Perfusion MRI
Perfusion MRI or perfusion-weighted imaging (PWI) is perfusion scanning by the use of a particular MRI sequence. The acquired data are then postprocessed to obtain perfusion maps with different parameters, such as BV (blood volume), BF (blood flow), MTT (mean transit time) and TTP (time to peak).
| Perfusion MRI | |
|---|---|
| Medical diagnostics | |
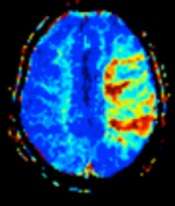 MRI perfusion showing a delayed time-to-maximum flow (Tmax) in the penumbra in a case of occlusion of the left middle cerebral artery. | |
| Purpose | perfusion scanning via MRI |
Clinical use
In cerebral infarction, the penumbra has decreased perfusion.[1] Another MRI sequence, diffusion weighted MRI, estimates the amount of tissue that is already necrotic, and the combination of those sequences can therefore be used to estimate the amount of brain tissue that is salvageable by thrombolysis and/or thrombectomy.[1]
Sequences
There are 3 main techniques for perfusion MRI:
- Dynamic susceptibility contrast (DSC): Gadolinium contrast is injected, and rapid repeated imaging (generally gradient-echo echo-planar T2 weighted) quantifies susceptibility-induced signal loss.[2]
- Dynamic contrast enhanced (DCE): Measuring shortening of the spin–lattice relaxation (T1) induced by a gadolinium contrast bolus[3]
- Arterial spin labelling (ASL): Magnetic labeling of arterial blood below the imaging slab, without the need of gadolinium contrast[4]
It can also be argued that diffusion MRI models, such as intravoxel incoherent motion, also attempt to capture perfusion.
Dynamic susceptibility contrast
In Dynamic susceptibility contrast MR imaging (DSC-MRI, or simply DSC), Gadolinium contrast agent (Gd) is injected (usually intravenously) and a time series of fast T2*-weighted images is acquired. As Gadolinium passes through the tissues, it induces a reduction of T2* in the nearby water protons; the corresponding decrease in signal intensity observed depends on the local Gd concentration, which may be considered a proxy for perfusion. The acquired time series data are then postprocessed to obtain perfusion maps with different parameters, such as BV (blood volume), BF (blood flow), MTT (mean transit time) and TTP (time to peak).
Dynamic contrast-enhanced imaging
Dynamic contrast-enhanced (DCE) imaging gives information about physiological tissue characteristics. For example, it enables analysis of blood vessels generated by a brain tumor. The contrast agent is blocked by the regular blood–brain barrier but not in the blood vessels generated by the tumor. The concentration of the contrast agent is measured as it passes from the blood vessels to the extracellular space of the tissue (it does not pass the membranes of cells) and as it goes back to the blood vessels.[5][6]
The contrast agents used for DCE-MRI are often gadolinium based. Interaction with the gadolinium (Gd) contrast agent (commonly a gadolinium ion chelate) causes the relaxation time of water protons to decrease, and therefore images acquired after gadolinium injection display higher signal in T1-weighted images indicating the present of the agent. It is important to note that, unlike some techniques such as PET imaging, the contrast agent is not imaged directly, but by an indirect effect on water protons. The common procedure for a DCE-MRI exam is to acquire a regular T1-weighted MRI scan (with no gadolinium), and then gadolinium is injected (usually as an intravenous bolus at a dose of 0.05–0.1 mmol/kg) before further T1-weighted scanning. DCE-MRI may be acquired with or without a pause for contrast injection and may have varying time resolution depending on preference – faster imaging (less than 10s per imaging volume) allows pharmacokinetic (PK) modelling of contrast agent but can limit possible image resolution. Slower time resolution allows more detailed images, but may limit interpretation to only looking at signal intensity curve shape. In general, persistent increased signal intensity (corresponding to decreased T1 and thus increased Gd interaction) in a DCE-MRI image voxel indicates permeable blood vessels characteristic of tumor tissue, where Gd has leaked into the extravascular extracellular space. In tissues with healthy cells or a high cell density, gadolinium re-enters the vessels faster since it cannot pass the cell membranes. In damaged tissues or tissues with a lower cell density, the gadolinium stays in the extracellular space longer.
Pharmacokinetic modelling of gadolinium in DCE-MRI is complex and requires choosing a model. There are a variety of models, which describe tissue structure differently, including size and structure of plasma fraction, extravascular extracellular space, and the resulting parameters relating to permeability, surface area, and transfer constants.[7] DCE-MRI can also provide model-independent parameters, such as T1 (which is not technically part of the contrast scan and can be acquired independently) and (initial) area under the gadolinium curve (IAUGC, often given with number of seconds from injection - i.e., IAUGC60), which may be more reproducible.[8] Accurate measurement of T1 is required for some pharmacokinetic models, which can be estimated from 2 pre-gadolinium images of varying excitation pulse flip angle,[9] though this method is not intrinsically quantitative.[10] Some models require knowledge of the arterial input function, which may be measured on a per patient basis or taken as a population function from literature, and can be an important variable for modelling.[11]
Arterial spin labelling
Arterial spin labelling (ASL) has the advantage of not relying on an injected contrast agent, instead inferring perfusion from a drop in signal observed in the imaging slice arising from inflowing spins (outside the imaging slice) having been selectively saturated. A number of ASL schemes are possible, the simplest being flow alternating inversion recovery (FAIR) which requires two acquisitions of identical parameters with the exception of the out-of-slice saturation; the difference in the two images is theoretically only from inflowing spins, and may be considered a 'perfusion map'.
References
- Chen, Feng (2012). "Magnetic resonance diffusion-perfusion mismatch in acute ischemic stroke: An update". World Journal of Radiology. 4 (3): 63–74. doi:10.4329/wjr.v4.i3.63. ISSN 1949-8470. PMC 3314930. PMID 22468186.
- Frank Gaillard; et al. "Dynamic susceptibility contrast (DSC) MR perfusion". Radiopaedia. Retrieved 2017-10-14.
- Frank Gaillard; et al. "Dynamic contrast enhanced (DCE) MR perfusion". Radiopaedia. Retrieved 2017-10-15.
- Frank Gaillard; et al. "Arterial spin labelling (ASL) MR perfusion". Radiopaedia. Retrieved 2017-10-15.
- Paul S. Tofts. "T1-weighted DCE Imaging Concepts: Modelling, Acquisition and Analysis" (PDF). paul-tofts-phd.org.uk. Retrieved 22 June 2013.
- Buckley, D.L., Sourbron, S.P. (2013). "Classic models for dynamic contrast enhanced MRI". NMR in Biomedicine. 26 (8): 1004–27. doi:10.1002/nbm.2940. PMID 23674304.
- Tofts, PS; Buckley, DL (1997). "Modeling tracer kinetics in dynamic Gd-DTPA MR imaging". Journal of Magnetic Resonance Imaging. 7 (1): 91–101. doi:10.1002/nbm.2940. PMID 9039598.
- Miyazaki, Keiko; Jerome, Neil P.; Collins, David J.; Orton, Matthew R.; d’Arcy, James A.; Wallace, Toni; Moreno, Lucas; Pearson, Andrew D. J.; Marshall, Lynley V.; Carceller, Fernando; Leach, Martin O.; Zacharoulis, Stergios; Koh, Dow-Mu (15 March 2015). "Demonstration of the reproducibility of free-breathing diffusion-weighted MRI and dynamic contrast enhanced MRI in children with solid tumours: a pilot study". European Radiology. 25 (9): 2641–50. doi:10.1007/s00330-015-3666-7. PMC 4529450. PMID 25773937.
- Fram, EK; Herfkens, RJ; Johnson, GA; Glover, GH; Karis, JP; Shimakawa, A; Perkins, TG; Pelc, NJ (1987). "Rapid calculation of T1 using variable flip angle gradient refocused imaging". Magnetic Resonance Imaging. 5 (3): 201–08. doi:10.1016/0730-725X(87)90021-X. PMID 3626789.
- Cheng, K; Koeck, PJ; Elmlund, H; Idakieva, K; Parvanova, K; Schwarz, H; Ternström, T; Hebert, H (2006). "Rapana thomasiana hemocyanin (RtH): comparison of the two isoforms, RtH1 and RtH2, at 19A and 16A resolution". Micron (Oxford, England : 1993). 37 (6): 566–76. doi:10.1016/j.micron.2005.11.014. PMID 16466927.
- Calamante, Fernando (October 2013). "Arterial input function in perfusion MRI: A comprehensive review". Progress in Nuclear Magnetic Resonance Spectroscopy. 74: 1–32. doi:10.1016/j.pnmrs.2013.04.002. PMID 24083460.