Developmental biology
Developmental biology is the study of the process by which animals and plants grow and develop. Developmental biology also encompasses the biology of regeneration, asexual reproduction, metamorphosis, and the growth and differentiation of stem cells in the adult organism.
In the late 20th century, the discipline largely transformed into evolutionary developmental biology.
Perspectives
The main processes involved in the embryonic development of animals are: regional specification, morphogenesis, cell differentiation, growth, and the overall control of timing explored in evolutionary developmental biology:
- Regional specification refers to the processes that create spatial pattern in a ball or sheet of initially similar cells. This generally involves the action of cytoplasmic determinants, located within parts of the fertilized egg, and of inductive signals emitted from signaling centers in the embryo. The early stages of regional specification do not generate functional differentiated cells, but cell populations committed to develop to a specific region or part of the organism. These are defined by the expression of specific combinations of transcription factors.
- Morphogenesis relates to the formation of three-dimensional shape. It mainly involves the orchestrated movements of cell sheets and of individual cells. Morphogenesis is important for creating the three germ layers of the early embryo (ectoderm, mesoderm and endoderm) and for building up complex structures during organ development.
- Cell differentiation relates specifically to the formation of functional cell types such as nerve, muscle, secretory epithelia etc. Differentiated cells contain large amounts of specific proteins associated with the cell function.
- Growth involves both an overall increase in size, and also the differential growth of parts (allometry) which contributes to morphogenesis. Growth mostly occurs through cell division but also through changes of cell size and the deposition of extracellular materials.
- The control of timing of events and the integration of the various processes with one another is the least well understood area of the subject. It remains unclear whether animal embryos contain a master clock mechanism or not.
The development of plants involves similar processes to that of animals. However plant cells are mostly immotile so morphogenesis is achieved by differential growth, without cell movements. Also, the inductive signals and the genes involved are different from those that control animal development.
Developmental processes
Cell differentiation
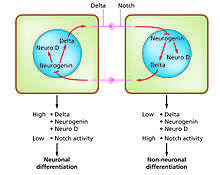
Cell differentiation is the process whereby different functional cell types arise in development. For example, neurons, muscle fibers and hepatocytes (liver cells) are well known types of differentiated cells. Differentiated cells usually produce large amounts of a few proteins that are required for their specific function and this gives them the characteristic appearance that enables them to be recognized under the light microscope. The genes encoding these proteins are highly active. Typically their chromatin structure is very open, allowing access for the transcription enzymes, and specific transcription factors bind to regulatory sequences in the DNA in order to activate gene expression.[1][2] For example, NeuroD is a key transcription factor for neuronal differentiation, myogenin for muscle differentiation, and HNF4 for hepatocyte differentiation. Cell differentiation is usually the final stage of development, preceded by several states of commitment which are not visibly differentiated. A single tissue, formed from a single type of progenitor cell or stem cell, often consists of several differentiated cell types. Control of their formation involves a process of lateral inhibition,[3] based on the properties of the Notch signaling pathway.[4] For example, in the neural plate of the embryo this system operates to generate a population of neuronal precursor cells in which NeuroD is highly expressed.
Regeneration
Regeneration indicates the ability to regrow a missing part.[5] This is very prevalent amongst plants, which show continuous growth, and also among colonial animals such as hydroids and ascidians. But most interest by developmental biologists has been shown in the regeneration of parts in free living animals. In particular four models have been the subject of much investigation. Two of these have the ability to regenerate whole bodies: Hydra, which can regenerate any part of the polyp from a small fragment,[6] and planarian worms, which can usually regenerate both heads and tails.[7] Both of these examples have continuous cell turnover fed by stem cells and, at least in planaria, at least some of the stem cells have been shown to be pluripotent.[8] The other two models show only distal regeneration of appendages. These are the insect appendages, usually the legs of hemimetabolous insects such as the cricket,[9] and the limbs of urodele amphibians.[10] Considerable information is now available about amphibian limb regeneration and it is known that each cell type regenerates itself, except for connective tissues where there is considerable interconversion between cartilage, dermis and tendons. In terms of the pattern of structures, this is controlled by a re-activation of signals active in the embryo. There is still debate about the old question of whether regeneration is a "pristine" or an "adaptive" property.[11] If the former is the case, with improved knowledge, we might expect to be able to improve regenerative ability in humans. If the latter, then each instance of regeneration is presumed to have arisen by natural selection in circumstances particular to the species, so no general rules would be expected.
Embryonic development of animals
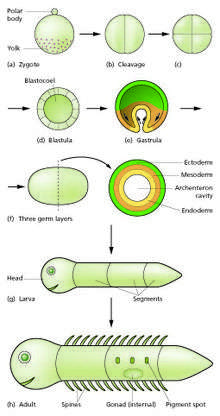
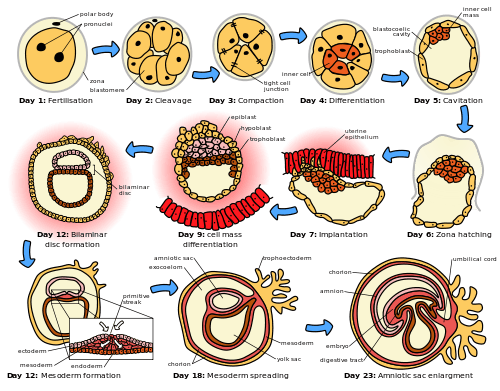
The sperm and egg fuse in the process of fertilization to form a fertilized egg, or zygote.[12] This undergoes a period of divisions to form a ball or sheet of similar cells called a blastula or blastoderm. These cell divisions are usually rapid with no growth so the daughter cells are half the size of the mother cell and the whole embryo stays about the same size. They are called cleavage divisions.
Mouse epiblast primordial germ cells (see Figure: “The initial stages of human embryogenesis”) undergo extensive epigenetic reprogramming.[13] This process involves genome-wide DNA demethylation, chromatin reorganization and epigenetic imprint erasure leading to totipotency.[13] DNA demethylation is carried out by a process that utilizes the DNA base excision repair pathway.[14]
Morphogenetic movements convert the cell mass into a three layered structure consisting of multicellular sheets called ectoderm, mesoderm and endoderm, which are known as germ layers. This is the process of gastrulation. During cleavage and gastrulation the first regional specification events occur. In addition to the formation of the three germ layers themselves, these often generate extraembryonic structures, such as the mammalian placenta, needed for support and nutrition of the embryo,[15] and also establish differences of commitment along the anteroposterior axis (head, trunk and tail).[16]
Regional specification is initiated by the presence of cytoplasmic determinants in one part of the zygote. The cells that contain the determinant become a signaling center and emit an inducing factor. Because the inducing factor is produced in one place, diffuses away, and decays, it forms a concentration gradient, high near the source cells and low further away.[17][18] The remaining cells of the embryo, which do not contain the determinant, are competent to respond to different concentrations by upregulating specific developmental control genes. This results in a series of zones becoming set up, arranged at progressively greater distance from the signaling center. In each zone a different combination of developmental control genes is upregulated.[19] These genes encode transcription factors which upregulate new combinations of gene activity in each region. Among other functions, these transcription factors control expression of genes conferring specific adhesive and motility properties on the cells in which they are active. Because of these different morphogenetic properties, the cells of each germ layer move to form sheets such that the ectoderm ends up on the outside, mesoderm in the middle, and endoderm on the inside.[20][21] Morphogenetic movements not only change the shape and structure of the embryo, but by bringing cell sheets into new spatial relationships they also make possible new phases of signaling and response between them.
Growth in embryos is mostly autonomous.[22] For each territory of cells the growth rate is controlled by the combination of genes that are active. Free-living embryos do not grow in mass as they have no external food supply. But embryos fed by a placenta or extraembryonic yolk supply can grow very fast, and changes to relative growth rate between parts in these organisms help to produce the final overall anatomy.
The whole process needs to be coordinated in time and how this is controlled is not understood. There may be a master clock able to communicate with all parts of the embryo that controls the course of events, or timing may depend simply on local causal sequences of events.[23]
Metamorphosis
Developmental processes are very evident during the process of metamorphosis. This occurs in various types of animal. Well-known are the examples of the frog, which usually hatches as a tadpole and metamorphoses to an adult frog, and certain insects which hatch as a larva and then become remodeled to the adult form during a pupal stage.
All the developmental processes listed above occur during metamorphosis. Examples that have been especially well studied include tail loss and other changes in the tadpole of the frog Xenopus,[24][25] and the biology of the imaginal discs, which generate the adult body parts of the fly Drosophila melanogaster.[26][27]
Plant development
Plant development is the process by which structures originate and mature as a plant grows. It is studied in plant anatomy and plant physiology as well as plant morphology.
Plants constantly produce new tissues and structures throughout their life from meristems[28] located at the tips of organs, or between mature tissues. Thus, a living plant always has embryonic tissues. By contrast, an animal embryo will very early produce all of the body parts that it will ever have in its life. When the animal is born (or hatches from its egg), it has all its body parts and from that point will only grow larger and more mature.
The properties of organization seen in a plant are emergent properties which are more than the sum of the individual parts. "The assembly of these tissues and functions into an integrated multicellular organism yields not only the characteristics of the separate parts and processes but also quite a new set of characteristics which would not have been predictable on the basis of examination of the separate parts."[29]
Growth
A vascular plant begins from a single celled zygote, formed by fertilisation of an egg cell by a sperm cell. From that point, it begins to divide to form a plant embryo through the process of embryogenesis. As this happens, the resulting cells will organize so that one end becomes the first root, while the other end forms the tip of the shoot. In seed plants, the embryo will develop one or more "seed leaves" (cotyledons). By the end of embryogenesis, the young plant will have all the parts necessary to begin in its life.
Once the embryo germinates from its seed or parent plant, it begins to produce additional organs (leaves, stems, and roots) through the process of organogenesis. New roots grow from root meristems located at the tip of the root, and new stems and leaves grow from shoot meristems located at the tip of the shoot.[30] Branching occurs when small clumps of cells left behind by the meristem, and which have not yet undergone cellular differentiation to form a specialized tissue, begin to grow as the tip of a new root or shoot. Growth from any such meristem at the tip of a root or shoot is termed primary growth and results in the lengthening of that root or shoot. Secondary growth results in widening of a root or shoot from divisions of cells in a cambium.[31]
In addition to growth by cell division, a plant may grow through cell elongation. This occurs when individual cells or groups of cells grow longer. Not all plant cells will grow to the same length. When cells on one side of a stem grow longer and faster than cells on the other side, the stem will bend to the side of the slower growing cells as a result. This directional growth can occur via a plant's response to a particular stimulus, such as light (phototropism), gravity (gravitropism), water, (hydrotropism), and physical contact (thigmotropism).
Plant growth and development are mediated by specific plant hormones and plant growth regulators (PGRs) (Ross et al. 1983).[32] Endogenous hormone levels are influenced by plant age, cold hardiness, dormancy, and other metabolic conditions; photoperiod, drought, temperature, and other external environmental conditions; and exogenous sources of PGRs, e.g., externally applied and of rhizospheric origin.
Morphological variation
Plants exhibit natural variation in their form and structure. While all organisms vary from individual to individual, plants exhibit an additional type of variation. Within a single individual, parts are repeated which may differ in form and structure from other similar parts. This variation is most easily seen in the leaves of a plant, though other organs such as stems and flowers may show similar variation. There are three primary causes of this variation: positional effects, environmental effects, and juvenility.
Evolution of plant morphology
Transcription factors and transcriptional regulatory networks play key roles in plant morphogenesis and their evolution. During plant landing, many novel transcription factor families emerged and are preferentially wired into the networks of multicellular development, reproduction, and organ development, contributing to more complex morphogenesis of land plants.[33]
Developmental model organisms
Much of developmental biology research in recent decades has focused on the use of a small number of model organisms. It has turned out that there is much conservation of developmental mechanisms across the animal kingdom. In early development different vertebrate species all use essentially the same inductive signals and the same genes encoding regional identity. Even invertebrates use a similar repertoire of signals and genes although the body parts formed are significantly different. Model organisms each have some particular experimental advantages which have enabled them to become popular among researchers. In one sense they are "models" for the whole animal kingdom, and in another sense they are "models" for human development, which is difficult to study directly for both ethical and practical reasons. Model organisms have been most useful for elucidating the broad nature of developmental mechanisms. The more detail is sought, the more they differ from each other and from humans.
Plants:
- Thale cress (Arabidopsis thaliana)
Vertebrates:
- Frog: Xenopus (X.laevis and tropicalis).[34][35] Good embryo supply. Especially suitable for microsurgery.
- Zebrafish: Danio rerio.[36] Good embryo supply. Well developed genetics.
- Chicken: Gallus gallus.[37] Early stages similar to mammal, but microsurgery easier. Low cost.
- Mouse: Mus musculus.[38] A mammal with well developed genetics.
Invertebrates:
- Fruit fly: Drosophila melanogaster.[39] Good embryo supply. Well developed genetics.
- Nematode: Caenorhabditis elegans.[40] Good embryo supply. Well developed genetics. Low cost.
Also popular for some purposes have been sea urchins[41] and ascidians.[42] For studies of regeneration urodele amphibians such as the axolotl Ambystoma mexicanum are used,[43] and also planarian worms such as Schmidtea mediterranea.[44] Organoids have also been demonstrated as an efficient model for development.[45] Plant development has focused on the thale cress Arabidopsis thaliana as a model organism.[46]
See also
- Blastocyst
- Body plan
- Cell signaling
- Cell signaling networks
- Embryology
- Enhancer
- Fish development
- Gene regulatory network
- Ontogeny
- Plant evolutionary developmental biology
- Promoter (biology)
- Signal transduction
- Teratology
References
- Li B.; Carey M.; Workman J.L. (2007). "The role of chromatin during transcription". Cell. 128 (4): 707–719. doi:10.1016/j.cell.2007.01.015. PMID 17320508.
- Heintzman N.D.; et al. (2007). "Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome". Nat Genet. 39 (3): 311–318. doi:10.1038/ng1966. PMID 17277777.
- Meinhardt H., Gierer A. (2000). "Pattern formation by local self-activation and lateral inhibition" (PDF). BioEssays. 22 (8): 753–760. CiteSeerX 10.1.1.477.439. doi:10.1002/1521-1878(200008)22:8<753::aid-bies9>3.0.co;2-z. Archived (PDF) from the original on 2017-10-27.
- Sprinzak D.; et al. (2010). "Cis-interactions between Notch and Delta generate mutually exclusive signalling states". Nature. 465 (7294): 86–90. doi:10.1038/nature08959. PMC 2886601. PMID 20418862.
- Carlson, B.M. (2007) Principles of Regenerative Biology. Academic Press, Burlington MA.
- Bosch T.C.G. (2007). "Why polyps regenerate and we don't: Towards a cellular and molecular framework for Hydra regeneration". Developmental Biology. 303 (2): 421–433. doi:10.1016/j.ydbio.2006.12.012. PMID 17234176.
- Reddien P.W., Alvarado A.S. (2004). "Fundamentals of planarian regeneration". Annual Review of Cell and Developmental Biology. 20: 725–757. doi:10.1146/annurev.cellbio.20.010403.095114. PMID 15473858.
- Wagner D.E.; Wang I.E.; Reddien P.W. (2011). "Clonogenic Neoblasts Are Pluripotent Adult Stem Cells That Underlie Planarian Regeneration". Science. 332 (6031): 811–816. doi:10.1126/science.1203983. PMC 3338249. PMID 21566185.
- Nakamura T.; et al. (2008). "Dissecting insect leg regeneration through RNA interference". Cellular and Molecular Life Sciences. 65 (1): 64–72. doi:10.1007/s00018-007-7432-0. PMID 18030418.
- Simon A., Tanaka E.M. (2013). "Limb regeneration". Wiley Interdisciplinary Reviews: Developmental Biology. 2 (2): 291–300. doi:10.1002/wdev.73. PMID 24009038.
- Slack, J.M.W. (2013) Essential Developmental Biology. Chapter 20. Wiley-Blackwell, Oxford.
- Jungnickel M.K.; Sutton K.A.; Florman H.M. (2003). "In the beginning: lessons from fertilization in mice and worms". Cell. 114 (4): 401–404. doi:10.1016/s0092-8674(03)00648-2. PMID 12941269.
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA (January 2013). "Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine". Science. 339 (6118): 448–52. doi:10.1126/science.1229277. PMC 3847602. PMID 23223451.
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA (July 2010). "Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway". Science. 329 (5987): 78–82. doi:10.1126/science.1187945. PMC 3863715. PMID 20595612.
- Steven, D.H. (ed.) (1975) Comparative Placentation. Academic Press, London
- Kimelman D., Martin B.L. (2012). "Anterior-posterior patterning in early development: three strategies". Wiley Interdisciplinary Reviews: Developmental Biology. 1 (2): 253–266. doi:10.1002/wdev.25. PMC 5560123. PMID 23801439.
- Slack J.M.W. (1987). "Morphogenetic gradients - past and present". Trends in Biochemical Sciences. 12: 200–204. doi:10.1016/0968-0004(87)90094-6.
- Rogers K. W., Schier A. F. (2011). "Morphogen Gradients: From Generation to Interpretation". Annual Review of Cell and Developmental Biology. 27: 377–407. doi:10.1146/annurev-cellbio-092910-154148.
- Dahmann C.; Oates A. C.; Brand M. (2011). "Boundary formation and maintenance in tissue development". Nat Rev Genet. 12: 43–55. doi:10.1038/nrg2902.
- Hardin J., Walston T. (2004). "Models of morphogenesis: the mechanisms and mechanics of cell rearrangement". Current Opinion in Genetics & Development. 14 (4): 399–406. doi:10.1016/j.gde.2004.06.008. PMID 15261656.
- Hammerschmidt M., Wedlich D. (2008). "Regulated adhesion as a driving force of gastrulation movements". Development. 135 (22): 3625–3641. doi:10.1242/dev.015701. PMID 18952908.
- O'Farrell, P. H. (2003). How metazoans reach their full size: the natural history of bigness. In Cell Growth: Control of Cell Size, (ed. M. N. Hall, Raff, M., and Thomas, G. (eds)), pp. 1-21: Cold Spring Harbor Laboratory Press
- Moss E.G., Romer-Seibert J. (2014). "Cell-intrinsic timing in animal development". Wiley Interdisciplinary Reviews: Developmental Biology. 3 (5): 365–377. doi:10.1002/wdev.145. PMID 25124757.
- Tata J.R. (1996). "Amphibian metamorphosis: an exquisite model for hormonal regulation of postembryonic development in vertebrates". Dev. Growth Diffn. 38 (3): 223–231. doi:10.1046/j.1440-169x.1996.t01-2-00001.x.
- Brown D.D., Cai L. (2007). "Amphibian metamorphosis". Developmental Biology. 306 (1): 20–33. doi:10.1016/j.ydbio.2007.03.021. PMC 1945045. PMID 17449026.
- Cohen, S.M. (1993) Imaginal Disc Development. In Bate and Martinez-Arias (eds.), The Development of Drosophila melanogaster, Cold Spring Harbor Press
- Maves L., Schubiger G. (2003). "Transdetermination in Drosophila imaginal discs: a model for understanding pluripotency and selector gene maintenance". Current Opinion in Genetics & Development. 13 (5): 472–479. doi:10.1016/j.gde.2003.08.006.
- Bäurle, I; Laux, T (2003). "Apical meristems: The plant's fountain of youth". BioEssays. 25 (10): 961–70. doi:10.1002/bies.10341. PMID 14505363. Review.
- Leopold, A. C. Plant Growth and Development, page 183. (New York: McGraw-Hill, 1964).
- Brand, U; Hobe, M; Simon, R (2001). "Functional domains in plant shoot meristems". BioEssays. 23 (2): 134–41. doi:10.1002/1521-1878(200102)23:2<134::AID-BIES1020>3.0.CO;2-3. PMID 11169586. Review.
- Barlow, P (2005). "Patterned cell determination in a plant tissue: The secondary phloem of trees". BioEssays. 27 (5): 533–41. doi:10.1002/bies.20214. PMID 15832381.
- Ross, S.D.; Pharis, R.P.; Binder, W.D. 1983. Growth regulators and conifers: their physiology and potential uses in forestry. p. 35–78 in Nickell, L.G. (Ed.), Plant growth regulating chemicals. Vol. 2, CRC Press, Boca Raton FL.
- Jin JP; et al. (July 2015). "An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors". Molecular Biology and Evolution. 32 (7): 1767–1773. doi:10.1093/molbev/msv058. PMC 4476157. PMID 25750178. Archived from the original on 2016-06-02.
- Nieuwkoop, P.D. and Faber, J. (1967) Normal table of Xenopus laevis (Daudin). North-Holland, Amsterdam.
- Harland R.M., Grainger R.M. (2011). "Xenopus research: metamorphosed by genetics and genomics". Trends in Genetics. 27 (12): 507–515. doi:10.1016/j.tig.2011.08.003. PMC 3601910. PMID 21963197.
- Lawson N. D., Wolfe S. A. (2011). "Forward and Reverse Genetic Approaches for the Analysis of Vertebrate Development in the Zebrafish". Developmental Cell. 21 (1): 48–64. doi:10.1016/j.devcel.2011.06.007. PMID 21763608.
- Hassan Rashidi V.S. (2009). "The chick embryo: hatching a model for contemporary biomedical research". BioEssays. 31 (4): 459–465. doi:10.1002/bies.200800168. PMID 19274658.
- Behringer, R., Gertsenstein, M, Vintersten, K. and Nagy, M. (2014) Manipulating the Mouse Embryo. A Laboratory Manual, Fourth Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- St Johnston D (2002). "The art and design of genetic screens: Drosophila melanogaster". Nat Rev Genet. 3 (3): 176–188. doi:10.1038/nrg751. PMID 11972155.
- Riddle, D.L., Blumenthal, T., Meyer, B.J. and Priess, J.R. (1997) C.elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ettensohn C.A., Sweet H.C. (2000). Patterning the early sea urchin embryo. Curr. Top. Dev. Biol. Current Topics in Developmental Biology. 50. pp. 1–44. doi:10.1016/S0070-2153(00)50002-7. ISBN 9780121531508. PMID 10948448.
- Lemaire P (2011). "Evolutionary crossroads in developmental biology: the tunicates". Development. 138 (11): 2143–2152. doi:10.1242/dev.048975. PMID 21558365.
- Nacu E., Tanaka E.M. (2011). "Limb Regeneration: A New Development?". Annual Review of Cell and Developmental Biology. 27: 409–440. doi:10.1146/annurev-cellbio-092910-154115. PMID 21801016.
- Reddien P.W., Alvarado A.S. (2004). "Fundamentals of planarian regeneration". Annual Review of Cell and Developmental Biology. 20: 725–757. doi:10.1146/annurev.cellbio.20.010403.095114. PMID 15473858.
- Ader M., Tanaka E. M. (2014). "Modeling human development in 3D culture". Current Opinion in Cell Biology. 31: 23–28. doi:10.1016/j.ceb.2014.06.013. PMID 25033469.
- Weigel, D. and Glazebrook, J. (2002) Arabidopsis. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Further reading
- Gilbert, S. F. (2013). Developmental Biology. Sunderland, Mass.: Sinauer Associates Inc.
- Slack, J. M. W. (2013). Essential Developmental Biology. Oxford: Wiley-Blackwell.
- Wolpert, L. and Tickle, C. (2011). Principles of Development. Oxford and New York: Oxford University Press.
External links
| Wikibooks has more on the topic of: Developmental biology |
| Wikimedia Commons has media related to Developmental biology. |