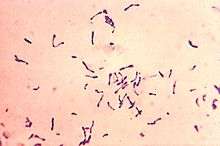
Corynebacterium diphtheriae
Corynebacterium diphtheriae[lower-alpha 1] is the pathogenic bacterium that causes diphtheria.[2] It is also known as the Klebs-Löffler bacillus, because it was discovered in 1884 by German bacteriologists Edwin Klebs (1834–1912) and Friedrich Löffler (1852–1915).
| Corynebacterium diphtheriae | |
|---|---|
 | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | C. diphtheriae |
| Binomial name | |
| Corynebacterium diphtheriae (Kruse 1886) Lehmann and Neumann 1896[1] | |
Classification
Four subspecies are recognized: C. d. mitis, C. d. intermedius, C. d. gravis, and C. d. belfanti. The four subspecies differ slightly in their colonial morphology and biochemical properties, such as the ability to metabolize certain nutrients, but all may be toxigenic (and therefore cause diphtheria) or not toxigenic. C. diphtheriae produces diphtheria toxin which alters protein function in the host by inactivating the elongation factor EF-2. This causes pharyngitis and 'pseudomembrane' in the throat. The diphtheria toxin gene is encoded by a bacteriophage found in toxigenic strains, integrated into the bacterial chromosome.
To accurately identify C. diphtheriae, a Gram stain is performed to show Gram-positive, highly pleomorphic organisms with no particular arrangement. Special stains like Albert’s stain and Ponder's stain are used to demonstrate the metachromatic granules formed in the polar regions. The granules are called polar granules, Babes Ernst granules, volutin, etc. An enrichment medium, such as Löffler's medium, is used to preferentially grow C. diphtheriae. After that, a differential plate known as tellurite agar, allows all Corynebacteria (including C. diphtheriae) to reduce tellurite to metallic tellurium. The tellurite reduction is colorimetrically indicated by brown colonies for most Cornyebacteria species or by a black halo around the C. diphtheriae colonies.
A low concentration of iron is required in the medium for toxin production. At high iron concentrations, iron molecules bind to an aporepressor on the beta bacteriophage, which carries the Tox gene. When bound to iron, the aporepressor shuts down toxin production.[3] Elek's test for toxigenicity is used to determine whether the organism is able to produce the diphtheria toxin.
Pathogen and disease
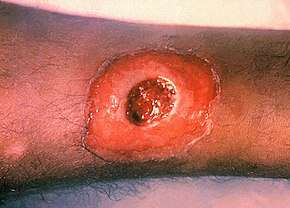
Corynebacterium diphtheriae is the bacterium that causes the disease diphtheria. Corynebacterium diphtheriae is a rod-shaped, Gram positive, non spore-forming, and nonmotile bacterium.[4] Although the geographic occurrence for the disease is worldwide, it is mainly to be found in tropical regions and underdeveloped countries. Those facing the greatest risk of contracting the disease are immunocompromised individuals, poorly immunized adults, and unvaccinated children. When an individual contracts diphtheria, the only affected region of the body is the upper respiratory system. Diphtheria can cause a thick gray coating to build up in the throat or nose making it difficult to breathe and swallow. Once individual contracts the disease, they are contagious for two weeks to a month.[5] The portals of entry for Corynebacterium diphtheriae are the nose, tonsils, and throat. Diseased individuals may experience a sore throat, overall weakness, fever, and swollen glands. Diphtheria is transmitted from human contact through respiratory droplets, such as coughing or sneezing. Although unlikely, individuals can get sick from touching open sores or clothes that touched open sores of someone sick with the disease. If untreated, the diphtheria toxin can get into the bloodstream causing damage to the kidneys, nerves, and heart. Extremely rare but serious complications include suffocation and partial paralysis. One preventive measure against diphtheria would be to get the DTap vaccine if available.
Pathogenesis
In areas where diphtheria is endemic, C. diphtheriae in the nasopharyngeal passageways is common. The exotoxin of Corynebacterium diptheriae is absorbed in the blood which in turn kills heart, kidney, and nerve cells by blocking protein synthesis.[6] Toxigenic strains in susceptible individuals can cause disease by multiplying and secreting diphtheria toxin into either skin or nasopharyngeal lesions. The diphtheritic lesion is often covered by a pseudomembrane composed of fibrin, bacteria, and inflammatory cells. Diphtheria toxin can be proteolytically cleaved into two fragments: an N-terminal fragment A (catalytic domain), and fragment B (transmembrane and receptor binding domain). Fragment A catalyzes the NAD+ -dependent ADP-ribosylation of elongation factor 2, thereby inhibiting protein synthesis in eukaryotic cells. Fragment B binds to the cell surface receptor and facilitates the delivery of fragment A to the cytosol.
Sensitivity
The bacterium is sensitive to the majority of antibiotics, such as the penicillins, ampicillin, cephalosporins, quinolones, chloramphenicol, tetracyclines, cefuroxime, and trimethoprim.
Genetics
The genome of C. diphtheriae consists of a single circular chromosome of 2.5 Mbp, with no plasmids.[7][8] The genome shows an extreme compositional bias, being noticeably higher in G+C near the origin than at the terminus.
See also
Notes
- Pronunciation: /kɔːˈraɪnəbæktɪəriəm dɪfˈθɪərii, -rɪnə-/.
References
- Parte, A.C. "Corynebacterium". www.bacterio.net.
- Hoskisson, P.A. (2018). "Microbe Profile: Corynebacterium diphtheriae – an old foe always ready to seize opportunity" (PDF). Microbiology. doi:10.1099/mic.0.000627. PMID 29465341.
- Nester, Eugene W.; et al. (2004). Microbiology: A Human Perspective (Fourth ed.). Boston: McGraw-Hill. ISBN 0-07-247382-7.
- "Diphtheria Infection | Home | CDC". www.cdc.gov. 2017-04-10. Retrieved 2017-11-27.
- "Diphtheria | MedlinePlus". Retrieved 2017-11-27.
- "Diphtheria". Healthline. Retrieved 2017-11-27.
- Cerdeño-Tárraga, A. M.; Efstratiou, A; Dover, L. G.; Holden, M. T.; Pallen, M; Bentley, S. D.; Besra, G. S.; Churcher, C; James, K. D.; De Zoysa, A; Chillingworth, T; Cronin, A; Dowd, L; Feltwell, T; Hamlin, N; Holroyd, S; Jagels, K; Moule, S; Quail, M. A.; Rabbinowitsch, E; Rutherford, K. M.; Thomson, N. R.; Unwin, L; Whitehead, S; Barrell, B. G.; Parkhill, J (2003). "The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129". Nucleic Acids Research. 31 (22): 6516–23. doi:10.1093/nar/gkg874. PMC 275568. PMID 14602910.
- Sangal, V; Tucker, N. P.; Burkovski, A; Hoskisson, P. A. (2012). "The draft genome sequence of Corynebacterium diphtheriae bv. Mitis NCTC 3529 reveals significant diversity between the primary disease-causing biovars". Journal of Bacteriology. 194 (12): 3269. doi:10.1128/JB.00503-12. PMC 3370853. PMID 22628502.
External links
- CoryneRegNet—Database of Corynebacterial Transcription Factors and Regulatory Networks
- Corynebacterium diphtheriae genome
- Type strain of Corynebacterium diphtheriae at BacDive - the Bacterial Diversity Metadatabase