Anti-sigma factors
In the regulation of gene expression in prokaryotes, anti-sigma factors bind to sigma factors and inhibit transcriptional activity. Anti-sigma factors have been found in a number of bacteria, including Escherichia coli and Salmonella, and in the T4 bacteriophage. Anti-sigma factors are antagonists to the sigma factors, which regulate numerous cell processes including flagellar production, stress response, transport and cellular growth. For example, anti-sigma factor 70 Rsd in E. coli is present in the stationary phase and blocks the activity of sigma factor 70 which in essence initiate gene transcription. This allows the sigma S factor to associate with RNA polymerase and direct the expression of the stationary genes. Although binding of Rsd to σ70 has been shown and numerous structural studies on Rsd have been performed, the detailed mechanism of action is still unknown.[1]
General information
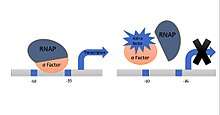
Sigma factor is an important protein which starts the transcription by binding with RNAP, anti-sigma factor is a protein which inhibits the activities of sigma factor affect by several mechanism such as add up anti-sigma factor between sigma or twist the anti-sigma factor around sigma.[2]
"In bacteria, the regulation of gene expression is the basis for adaptability, morphogenesis, and cellular differentiation. From all the different regulatory layers, regulation of transcription initiation is a very important step for controlling gene expression."[3]
As an example of a mechanism function of Anti-Sigma factors that it’s usually can play an important role instead of being a sigma factors destroyers since they inhibit some cell processes and activate another depending on some researches has been done. Researchers has study several anti-sigma factor activities in the cell such as RsrA (Redox-sensitive), which is a key sensor of thiol–disulphide and a negative regulator ofσR (binds σR and inhibits σR-directed transcription only in vitro), what means that it needs some sensitive conditions to happen. They conclude by using Thioredoxin that it’s able to reduce the oxidize RsrA.
Mechanism
There are four steps to start the complex: firstly, the relative sigma factor be appeared in particular environmental signal. Secondly, different transcriptional factor and sigma factor interact with their correspondent anti sigma factor. Thirdly, anti-sigma factor has different roles: either stay not active or activate it in specific environmental signal. Finally, since the signals disappear, anti-sigma factor inhibit the sigma factor by confinement.[2]
Specialized anti-sigma factors
In prokaryotes, E. coli has seven different sigma factors depends on the environment condition. Each one specific anti-sigma factors,[4] check the below table for details:
| Sigma factor | Related anti-sigma factor |
| σ70 | Rsd and HscC |
| σ24 | RseA |
| σ28 | FIgM |
| σ32 | Dnak |
| σ38 | RssB |
| σ54 | EBPs |
| σ19 | FecR |
As another Anti-sigma factor example is σH which regulate the somatic stress response and the morphological differentiation in S treptomyces coelicolor as the studies results. From the same study it can be concluded that every anti-sigma factor has a specific gene location to act and affect in the cell.
Anti sigma factor and E-coli
As a negative feedback interaction, anti-sigma factor in E-coli, help the cell or allow it to control the gene expression in a particular environment terms. As soon as these terms disappeared, the gene expression complete the circle.
In Bacteriophage
T4 bacteriophage uses anti- sigma factor to ruin the Escherichia coli polymerase in order that direct exclusive transcription of its own genes.
(AsiA is an anti-sigma factor gene that it’s required for bacteriophage T4 to be developed). What means that AsiA is an essential anti sigma factor in bacteriophage.
References
- Hofmann N, Wurm R, Wagner R (May 2011). "The E. coli anti-sigma factor Rsd: studies on the specificity and regulation of its expression". PLOS One. 6 (5): e19235. doi:10.1371/journal.pone.0019235. PMC 3089606. PMID 21573101.
- Paget MS (June 2015). "Bacterial Sigma Factors and Anti-Sigma Factors: Structure, Function and Distribution". Biomolecules. 5 (3): 1245–65. doi:10.3390/biom5031245. PMC 4598750. PMID 26131973.
- Hughes KT, Mathee K (1998). "The anti-sigma factors". Annual Review of Microbiology. 52: 231–86. doi:10.1146/annurev.micro.52.1.231. PMID 9891799.
- Treviño-Quintanilla LG, Freyre-González JA, Martínez-Flores I (September 2013). "Anti-Sigma Factors in E. coli: Common Regulatory Mechanisms Controlling Sigma Factors Availability". Current Genomics. 14 (6): 378–87. doi:10.2174/1389202911314060007. PMC 3861889. PMID 24396271.
5- [1]
6- [2]
7- [3]
Further reading
- Kang JG, Paget MS, Seok YJ, Hahn MY, Bae JB, Hahn JS, Kleanthous C, Buttner MJ, Roe JH (August 1999). "RsrA, an anti-sigma factor regulated by redox change". The EMBO Journal. 18 (15): 4292–8. doi:10.1093/emboj/18.15.4292. PMC 1171505. PMID 10428967.
- Ohnishi K, Kutsukake K, Suzuki H, Lino T (November 1992). "A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an antisigma factor inhibits the activity of the flagellum-specific sigma factor, sigma F". Molecular Microbiology. 6 (21): 3149–57. doi:10.1111/j.1365-2958.1992.tb01771.x. PMID 1453955.
- Helmann JD (April 1999). "Anti-sigma factors". Current Opinion in Microbiology. 2 (2): 135–41. doi:10.1016/S1369-5274(99)80024-1. PMID 10322161.
- Sevcikova B, Rezuchova B, Homerova D, Kormanec J (November 2010). "The anti-anti-sigma factor BldG is involved in activation of the stress response sigma factor σ(H) in Streptomyces coelicolor A3(2)". Journal of Bacteriology. 192 (21): 5674–81. doi:10.1128/JB.00828-10. PMC 2953704. PMID 20817765.
- Kang, Ju-Gyeong; Paget, Mark S. B.; Seok, Yeong-Jae; Hahn, Mi-Young; Bae, Jae-Bum; Hahn, Ji-Sook; Kleanthous, Colin; Buttner, Mark J.; Roe, Jung-Hye (1999-08-02). "RsrA, an anti‐sigma factor regulated by redox change". The EMBO Journal. 18 (15): 4292–4298. doi:10.1093/emboj/18.15.4292. ISSN 0261-4189. PMC 1171505. PMID 10428967.
- Treviño-Quintanilla, Luis Gerardo; Freyre-González, Julio Augusto; Martínez-Flores, Irma (September 2013). "Anti-Sigma Factors in E. coli: Common Regulatory Mechanisms Controlling Sigma Factors Availability". Current Genomics. 14 (6): 378–387. doi:10.2174/1389202911314060007. ISSN 1389-2029. PMC 3861889. PMID 24396271.
- Paget, Mark S. B.; Bae, Jae-Bum; Hahn, Mi-Young; Li, Wei; Kleanthous, Colin; Roe, Jung-Hye; Buttner, Mark J. (2001-12-21). "Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch". Molecular Microbiology. 39 (4): 1036–1047. doi:10.1046/j.1365-2958.2001.02298.x. ISSN 0950-382X.